Exploring the Potential Effects and Mechanisms of Asarum sieboldii Radix Essential Oil for Treatment of Asthma
Abstract
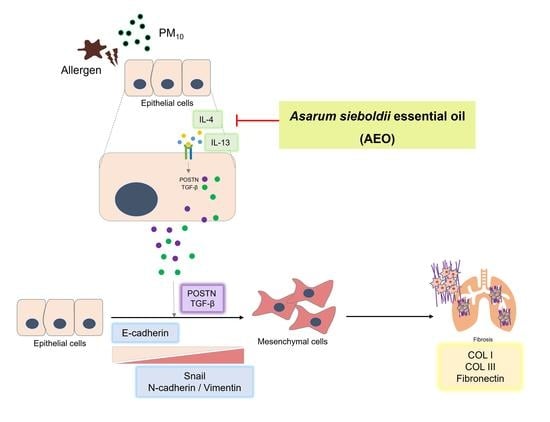
:1. Introduction
2. Materials and Methods
2.1. Network Pharmacological Analysis
2.2. Preparation of AEO
2.3. Animal Treatment
2.4. Histological Analysis
2.5. Cell Counting in BALF
2.6. Enzyme-Linked Immunosorbent Assay
2.7. Cell Treatment
2.8. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.9. Protein Immunoblot Analysis
2.10. Statistical Analysis
3. Results
3.1. Gene Comparison with AEO and Asthma
3.2. Histological Structure of Lung and Trachea Tissues in OVA+PM10-Induced Mice
3.3. Goblet Cell Accumulation in Lung and Trachea Tissues in OVA+PM10-Induced Mice
3.4. Collagen Deposition of Lung Tissues in OVA+PM10-Induced Mice
3.5. Inflammatory Cell Counts in BALF of OVA+PM10-Induced Mice
3.6. Serum IgE and IgG2a in OVA+PM10-Induced Mice
3.7. Expressions of Pro-Inflammatory and Th2 Cytokines in OVA+PM10-Induced Lung Tissues
3.8. Expressions of MMPs in OVA+PM10-Induced Lung Tissues
3.9. Expressions of Fibrotic Mediators in OVA+PM10-Induced Lung Tissues and PM10-Induced A549 Cells
3.10. Expressions of Epithelial–Mesenchymal Transition (EMT) Markers in OVA+PM10-Induced Lung Tissues and PM10-Induced A549 Cells
3.11. The Predicted Functional Partners of POSTN by STRING Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- India State-Level Disease Burden Initiative Air Pollution Collaborators. Health and economic impact of air pollution in the states of India: The Global Burden of Disease Study 2019. Lancet Planet Health 2021, 5, e25–e38. [Google Scholar] [CrossRef]
- Castaneda, A.R.; Bein, K.J.; Smiley-Jewell, S.; Pinkerton, K.E. Fine particulate matter (PM2.5) enhances allergic sensitization in BALB/c mice. J. Toxicol. Environ. Health A 2017, 80, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, J.R.; Lloyd, C.M. Chronic inflammation and asthma. Mutat. Res. 2010, 690, 24–39. [Google Scholar] [CrossRef] [PubMed]
- Dharmage, S.C.; Perret, J.L.; Custovic, A. Epidemiology of asthma in children and adults. Front. Pediatr. 2019, 7, 246. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.K.; Shadie, A.M.; Bucknall, M.P.; Rutlidge, H.; Garthwaite, L.; Herbert, C.; Halliburton, B.; Parsons, K.S.; Wark, P.A. Differential injurious effects of ambient and traffic-derived particulate matter on airway epithelial cells. Respirology 2015, 20, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Zhang, J.; Yang, X.; Zhang, Y.; Chen, Z. The role and potential pathogenic mechanism of particulate matter in childhood asthma: A review and perspective. J. Immunol. Res. 2020, 2020, 8254909. [Google Scholar] [CrossRef] [Green Version]
- Moldoveanu, B.; Otmishi, P.; Jani, P.; Walker, J.; Sarmiento, X.; Guardiola, J.; Saad, M.; Yu, J. Inflammatory mechanisms in the lung. J. Inflamm. Res. 2009, 2, 1–11. [Google Scholar]
- Vignola, A.M.; Mirabella, F.; Costanzo, G.; Di Giorgi, R.; Gjomarkaj, M.; Bellia, V.; Bonsignore, G. Airway remodeling in asthma. Chest 2003, 123, 417S–422S. [Google Scholar] [CrossRef]
- Busse, W.W.; Kraft, M. Current unmet needs and potential solutions to uncontrolled asthma. Eur. Respir. Rev. 2022, 31, 163. [Google Scholar] [CrossRef]
- Caimmi, D.; Neukirch, C.; Louis, R.; Malard, O.; Thabut, G.; Demoly, P. Effect of the use of intranasal spray of essential oils in patients with perennial allergic rhinitis: A prospective study. Int. Arch. Allergy Immunol. 2021, 182, 182–189. [Google Scholar] [CrossRef]
- Ueno-Iio, T.; Shibakura, M.; Yokota, K.; Aoe, M.; Hyoda, T.; Shinohata, R.; Kanehiro, A.; Tanimoto, M.; Kataoka, M. Lavender essential oil inhalation suppresses allergic airway inflammation and mucous cell hyperplasia in a murine model of asthma. Life Sci. 2014, 108, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Mohammadhosseini, M.; Akbarzadeh, A.; Flamini, G. Profiling of compositions of essential oils and volatiles of Salvia limbata using traditional and advanced techniques and evaluation for biological activities of their extracts. Chem. Biodivers. 2017, 14, e1600361. [Google Scholar] [CrossRef] [PubMed]
- Mohammadhosseini, M.; Sarker, S.D.; Akbarzadeh, A. Chemical composition of the essential oils and extracts of Achillea species and their biological activities: A review. J. Ethnopharmacol. 2017, 199, 257–315. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.M.; Kim, J.; Lee, J.; Yi, J.M.; Oh, D.S.; Bang, O.S.; Kim, N.S. Anticancer potential of an ethanol extract of Asiasari radix against HCT-116 human colon cancer cells in vitro. Oncol. Lett. 2013, 5, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Kim, S.J. Memory enhancing actions of Asiasari radix extracts via activation of insulin receptor and extracellular signal regulated kinase (ERK) I/II in rat hippocampus. Brain Res. 2003, 974, 193–201. [Google Scholar] [CrossRef]
- Jang, J.Y.; Lee, J.H.; Shin, H.K.; Choi, Y.H.; Lee, J.D.; Choi, B.T. Partially purified Asiasari radix inhibits melanogenesis through extracellular signal-regulated kinase signaling in B16F10 cells. Int. J. Mol. Med. 2010, 25, 287–292. [Google Scholar]
- Kim, H.M.; Moon, Y.S. Asiasari radix inhibits immunoglobulin E production on experimental models in vitro and in vivo. Immunopharmacol. Immunotoxicol. 1999, 21, 469–481. [Google Scholar] [CrossRef]
- Wu, H.; Li, J.; Zhang, F.; Li, L.; Liu, Z.; He, Z. Essential oil components from Asarum sieboldii Miquel are toxic to the house dust mite Dermatophagoides farinae. Parasitol. Res. 2012, 111, 1895–1899. [Google Scholar] [CrossRef]
- Zhang, Z.; Kang, H. Protective effect of Asarum sieboldii essential oil on ovalbumin induced allergic rhinitis in rat. Biosci. Rep. 2020, 40, BSR20191370. [Google Scholar] [CrossRef]
- Tan, K.H.; Nishida, R. Methyl eugenol: Its occurrence, distribution, and role in nature, especially in relation to insect behavior and pollination. J. Insect Sci. 2012, 12, 56. [Google Scholar] [CrossRef] [Green Version]
- Southwell, I.A.; Russellb, M.F.; Davies, N.W. Detecting traces of methyl eugenol in essential oils: Tea tree oil, a case study. Flavour Fragr. J. 2011, 26, 336–340. [Google Scholar] [CrossRef]
- Gu, S.S.; Zhang, Y.; Li, X.L.; Wu, S.Y.; Diao, T.Y.; Hai, R.; Deng, H.W. Involvement of the skeletal renin-angiotensin system in age-related osteoporosis of ageing mice. Biosci. Biotechnol. Biochem. 2012, 76, 1367–1371. [Google Scholar] [CrossRef] [PubMed]
- Leong, K.P.; Huston, D.P. Understanding the pathogenesis of allergic asthma using mouse models. Ann. Allergy Asthma Immunol. 2001, 87, 96–109. [Google Scholar] [CrossRef]
- Falcon-Rodriguez, C.I.; Osornio-Vargas, A.R.; Sada-Ovalle, I.; Segura-Medina, P. Aeroparticles, composition, and lung diseases. Front. Immunol. 2016, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Park, E.J.; Yoon, C.; Han, J.S.; Lee, G.H.; Kim, D.W.; Park, E.J.; Lim, H.J.; Kang, M.S.; Han, H.Y.; Seol, H.J.; et al. Effect of PM10 on pulmonary immune response and fetus development. Toxicol. Lett. 2021, 339, 1–11. [Google Scholar] [CrossRef]
- Bergeron, C.; Tulic, M.K.; Hamid, Q. Airway remodelling in asthma: From benchside to clinical practice. Can. Respir. J. 2010, 17, e85–e93. [Google Scholar] [CrossRef] [Green Version]
- Iwanaga, K.; Elliott, M.S.; Vedal, S.; Debley, J.S. Urban particulate matter induces pro-remodeling factors by airway epithelial cells from healthy and asthmatic children. Inhal. Toxicol. 2013, 25, 653–660. [Google Scholar] [CrossRef]
- Bajbouj, K.; Ramakrishnan, R.K.; Hamid, Q. Role of matrix metalloproteinases in angiogenesis and its implications in asthma. J. Immunol. Res. 2021, 2021, 6645072. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Patel, K.D. Matrix metalloproteinase-2 (MMP-2) and MMP-9 in pulmonary pathology. Exp. Lung Res. 2005, 31, 599–621. [Google Scholar] [CrossRef]
- Kurai, J.; Watanabe, M.; Sano, H.; Hantan, D.; Shimizu, E. The effect of seasonal variations in airborne particulate matter on asthma-related airway inflammation in mice. Int. J. Environ. Res. Public Health 2016, 13, 579. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, S.G.; Lloyd, C.M. Eosinophils in the pathogenesis of allergic airways disease. Cell. Mol. Life Sci. 2007, 64, 1269–1289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiraiwa, K.; van Eeden, S.F. Contribution of lung macrophages to the inflammatory responses induced by exposure to air pollutants. Mediat. Inflamm. 2013, 2013, 619523. [Google Scholar] [CrossRef] [Green Version]
- Makinde, T.; Murphy, R.F.; Agrawal, D.K. The regulatory role of TGF-beta in airway remodeling in asthma. Immunol. Cell Biol. 2007, 85, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Tang, Y.; Xu, J. Effects of indoor coal fine particulate matter on the expression levels of inflammatory factors in ovalbumin-induced mice. Toxicol. Res. 2019, 8, 57–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nygaard, U.C.; Aase, A.; Lovik, M. The allergy adjuvant effect of particles—Genetic factors influence antibody and cytokine responses. BMC Immunol. 2005, 6, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, J.W.; Tjota, M.Y.; Sperling, A.I. The contribution of allergen-specific IgG to the development of th2-mediated airway inflammation. J. Allergy 2012, 2012, 236075. [Google Scholar] [CrossRef] [Green Version]
- Todd, N.W.; Luzina, I.G.; Atamas, S.P. Molecular and cellular mechanisms of pulmonary fibrosis. Fibrogenes. Tissue Repair 2012, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Suki, B.; Bates, J.H. Extracellular matrix mechanics in lung parenchymal diseases. Respir. Physiol. Neurobiol. 2008, 163, 33–43. [Google Scholar] [CrossRef] [Green Version]
- O’Dwyer, D.N.; Moore, B.B. The role of periostin in lung fibrosis and airway remodeling. Cell. Mol. Life Sci. 2017, 74, 4305–4314. [Google Scholar] [CrossRef]
- Izuhara, K.; Matsumoto, H.; Ohta, S.; Ono, J.; Arima, K.; Ogawa, M. Recent developments regarding periostin in bronchial asthma. Allergol. Int. 2015, 64 (Suppl. 1), S3–S10. [Google Scholar] [CrossRef] [Green Version]
- Harris, W.T.; Kelly, D.R.; Zhou, Y.; Wang, D.; MacEwen, M.; Hagood, J.S.; Clancy, J.P.; Ambalavanan, N.; Sorscher, E.J. Myofibroblast differentiation and enhanced TGF-B signaling in cystic fibrosis lung disease. PLoS ONE 2013, 8, e70196. [Google Scholar] [CrossRef]
- Sidhu, S.S.; Yuan, S.; Innes, A.L.; Kerr, S.; Woodruff, P.G.; Hou, L.; Muller, S.J.; Fahy, J.V. Roles of epithelial cell-derived periostin in TGF-beta activation, collagen production, and collagen gel elasticity in asthma. Proc. Natl. Acad. Sci. USA 2010, 107, 14170–14175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horowitz, J.C.; Thannickal, V.J. Epithelial-mesenchymal interactions in pulmonary fibrosis. Semin. Respir. Crit. Care Med. 2006, 27, 600–612. [Google Scholar] [CrossRef] [Green Version]
- Rout-Pitt, N.; Farrow, N.; Parsons, D.; Donnelley, M. Epithelial mesenchymal transition (EMT): A universal process in lung diseases with implications for cystic fibrosis pathophysiology. Respir. Res. 2018, 19, 136. [Google Scholar] [CrossRef]
- Bartis, D.; Mise, N.; Mahida, R.Y.; Eickelberg, O.; Thickett, D.R. Epithelial-mesenchymal transition in lung development and disease: Does it exist and is it important? Thorax 2014, 69, 760–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.M.; Kim, M.H.; Choi, L.Y.; Kim, G.; Yang, W.M. Exploring the Potential Effects and Mechanisms of Asarum sieboldii Radix Essential Oil for Treatment of Asthma. Pharmaceutics 2022, 14, 558. https://doi.org/10.3390/pharmaceutics14030558
Han JM, Kim MH, Choi LY, Kim G, Yang WM. Exploring the Potential Effects and Mechanisms of Asarum sieboldii Radix Essential Oil for Treatment of Asthma. Pharmaceutics. 2022; 14(3):558. https://doi.org/10.3390/pharmaceutics14030558
Chicago/Turabian StyleHan, Jae Min, Mi Hye Kim, La Yoon Choi, Gyeongsang Kim, and Woong Mo Yang. 2022. "Exploring the Potential Effects and Mechanisms of Asarum sieboldii Radix Essential Oil for Treatment of Asthma" Pharmaceutics 14, no. 3: 558. https://doi.org/10.3390/pharmaceutics14030558