Simple and Fast DNA Based Sensor System for Screening of Small-Molecule Compounds Targeting Eukaryotic Topoisomerase 1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. DNA Oligonucleotides
- 5′-amine REEAD primer: 5′-amine-CCAACCAACCAACCAAATAAG CGATCTTCACAGT- 3′;
- REEAD dumbbell substrate: 5′-AGAAAAATTTTTAAAAAAACTGTGAAGATC GCTTATTTTT TTAAAAATTT TTCTAAGTCT TTTA GATCCC TCAATGCTGC TGCTGTACTA CGATCTAAAA GACTTAGA-3′-amine;
- REEAD probe: 5′-FAM- CCTCAATGCT GCTGCT GTACTAC-3′.
- 5′-amine REEAD (C|L) primer: 5′-amine-CCAACCAACCAACCAAGGAGCCAAACATGTGCATTGAGG-3′;
- Cleavage half-dumbbell: 5′-phospho-AAA AAT TTT TTC TAA GTC TTT TAC CCT CAA TGC ACA TGT TTG GCT CCG TAA AAG ACT TAG A-3′-amine;
- Ligator half-dumbbell: 5′-AGA AAA AAT TTT TAG CTC GAA CTG TGA AGA TCG CTT ATT CGA GCT-3′;
- REEAD(C|L) probe 5′-FAM ACTGTGAAGATCGCTTAT-3′.
2.3. Phi29 Polymerase Purification
2.4. REEAD-on-a-Slide
2.5. T4 DNA-Ligase Purification
2.6. REEAD (C|L)
2.7. Colorimetric REEAD-on-a-Slide Using Silver-on-Gold Precipitation
2.8. Statistic
3. Results
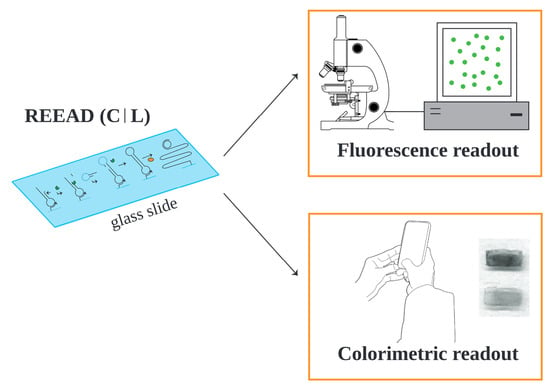
3.1. REEAD-on-a-Slide and REEAD (C|L) Assay Setups
3.2. REEAD (C|L) Can Be Used to Investigate TOP1 Binding/Cleavage and Ligation Separately
3.3. Measurement of TOP1 Inhibition by Small-Molecule Compounds Using REEAD (C|L)
3.4. Signal Readout Using Silver-on-Gold Precipitation
4. Discussion and Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Balaña-Fouce, R.; Redondo, C.M.; Pérez-Pertejo, Y.; Díaz-González, R.; Reguera, R.M. Targeting atypical trypanosomatid DNA topoisomerase I. Drug Discov. Today 2006, 11, 733–740. [Google Scholar] [CrossRef]
- García-Estrada, C.; Prada, C.F.; Fernández-Rubio, C.; Rojo-Vázquez, F.; Balaña-Fouce, R. DNA topoisomerases in apicomplexan parasites: Promising targets for drug discovery. Proc. R. Soc. B Biol. Sci. 2010, 277, 1777–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pommier, Y. Camptothecins and Topoisomerase I: A Foot in the Door. Targeting the genome beyond Topoisomerase I with camptothecins and Cycle Checkpoints. Curr. Med. Chem. Anticancer Agents 2004, 4, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, O.K.; Zuma, A.A.; Da Silva, C.C.; De Souza, W.; Motta, M.C.M. Effects of camptothecin derivatives and topoisomerase dual inhibitors on Trypanosoma cruzi growth and ultrastructure. J. Negat. Results Biomed. 2014, 13, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Álvarez-Bardón, M.; Pérez-Pertejo, Y.; Ordóñez, C.; Sepúlveda-Crespo, D.; Carballeira, N.M.; Tekwani, B.L.; Murugesan, S.; Martinez-Valladares, M.; García-Estrada, C.; Reguera, R.M.; et al. Screening marine natural products for new drug leads against trypanosomatids and malaria. Mar. Drugs 2020, 18, 187. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Sun, Y.; Huang, S.N.; Nitiss, J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016, 17, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Champoux, J.J. DNA TOPOISOMERASES: Structure, Function, and Mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. [Google Scholar] [CrossRef] [Green Version]
- Pommier, Y. (Ed.) DNA Topoisomerases and Cancer; Cancer Drug Discovery and Development Series; Springer: New York, NY, USA, 2012. [Google Scholar]
- Thomas, A.; Pommier, Y. Targeting topoisomerase I in the era of precision medicine. Clin. Cancer Res. 2019, 25, 6581–6589. [Google Scholar] [CrossRef]
- Cinelli, M.A. Topoisomerase 1B poisons: Over a half-century of drug leads, clinical candidates, and serendipitous discoveries. Med. Res. Rev. 2019, 39, 1294–1337. [Google Scholar] [CrossRef]
- Stewart, L.; Redinbo, M.R.; Qiu, X.; Hol, W.G.; Champoux, J.J. A model for the mechanism of human topoisomerase I. Science 1998, 279, 1534–1541. [Google Scholar] [CrossRef]
- Balaña-Fouce, R.; García-Estrada, C.; Pérez-Pertejo, Y.; Reguera, R.M. Gene disruption of the DNA topoisomerase IB small subunit induces a non-viable phenotype in the hemoflagellate Leishmania major. BMC Microbiol. 2008, 8, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakshi, R.P.; Shapiro, T.A. RNA interference of Trypanosoma brucei topoisomerase IB: Both subunits are essential. Mol. Biochem. Parasitol. 2004, 136, 249–255. [Google Scholar] [CrossRef]
- Zhang, C.X.; Chen, A.D.; Gettel, N.J.; Hsieh, T.S. Essential functions of DNA topoisomerase I in Drosophila melanogaster. Dev. Biol. 2000, 222, 27–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morham, S.G.; Kluckman, K.D.; Voulomanos, N.; Smithies, O. Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol. Cell. Biol. 1996, 16, 6804–6809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitiss, J.L.; Soans, E.; Rogojina, A.; Seth, A.; Mishina, M. Topoisomerase assays. In Current Protocols in Pharmacology; Wiley Press: Hoboken, NJ, USA, 2012; pp. 789–802. ISBN 9780896034440. [Google Scholar]
- Tesauro, C.; Fiorani, P.; D’Annessa, I.; Chillemi, G.; Turchi, G.; Desideri, A. Erybraedin C, a natural compound from the plant Bituminaria bituminosa, inhibits both the cleavage and religation activities of human topoisomerase I. Biochem. J. 2010, 425, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Christiansen, K.; Westergaards, O. Characterization of Intra- and Intermolecular DNA Ligation Mediated by Eukaryotic Topoisomerase. J. Biol. Chem. 1994, 269, 721–729. [Google Scholar] [CrossRef]
- Castelli, S.; Coletta, A.; D’Annessa, I.; Fiorani, P.; Tesauro, C.; Desideri, A. Interaction between natural compounds and human topoisomerase I. Biol. Chem. 2012, 393, 1327–1340. [Google Scholar] [CrossRef]
- Svejstrup, J.Q.; Christiansen, K.; Andersen, A.H.; Lund, K.; Westergaard, O. Minimal DNA duplex requirements for topoisomerase I-mediated cleavage in vitro. J. Biol. Chem. 1990, 265, 12529–12535. [Google Scholar] [CrossRef]
- Svejstrup, J.Q.; Christiansen, K.; Gromovat, I.I.; Andersen, A.H. New Technique for Uncoupling the Cleavage and Religation Reactions of Eukaryotic Topoisomerase I. The Mode of Action of Camptothecin at a Specific Recognition Site. J. Mol. Biol. 1991, 222, 669–678. [Google Scholar] [CrossRef]
- Stougaard, M.; Lohmann, J.S.; Mancino, A.; Celik, S.; Andersen, F.F.; Koch, J.; Knudsen, B.R. Single-molecule detection of human topoisomerase I cleavage-ligation activity. ACS Nano 2009, 3, 223–233. [Google Scholar] [CrossRef]
- Andersen, F.F.; Stougaard, M.; Jørgensen, H.L.; Bendsen, S.; Juul, S.; Hald, K.; Andersen, A.H.; Koch, J.; Knudsen, B.R. Multiplexed detection of site specific recombinase and DNA topoisomerase activities at the single molecule level. ACS Nano 2009, 3, 4043–4054. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.G.; Tesauro, C.; Coletta, A.; Graversen, A.D.; Ho, Y.-P.; Kristensen, P.; Stougaard, M.; Knudsen, B.R. On-slide detection of enzymatic activities in selected single cells. Nanoscale 2017, 9, 13546–13553. [Google Scholar] [CrossRef]
- Proszek, J.; Roy, A.; Jakobsen, A.K.; Frøhlich, R.; Knudsen, B.R.; Stougaard, M. Topoisomerase I as a biomarker: Detection of activity at the single molecule level. Sensors 2014, 14, 1195–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tesauro, C.; Keller, J.G.; Gromova, I.; Gromov, P.; Frøhlich, R.; Erlandsen, J.U.; Andersen, A.H.; Stougaard, M.; Knudsen, B.R. Different Camptothecin Sensitivities in Subpopulations of Colon Cancer Cells Correlate with Expression of Different Phospho-Isoforms of Topoisomerase I with Different Activities. Cancers 2020, 12, 1240. [Google Scholar] [CrossRef]
- Kjeldsen, E.; Nielsen, C.J.F.; Roy, A.; Tesauro, C.; Jakobsen, A.-K.; Stougaard, M.; Knudsen, B.R. Characterization of Camptothecin-induced Genomic Changes in the Camptothecin-resistant T-ALL-derived Cell Line CPT-K5. Cancer Genom. Proteom. 2018, 15, 91–114. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Tesauro, C.; Frøhlich, R.; Hede, M.S.; Nielsen, M.J.; Kjeldsen, E.; Bonven, B.; Stougaard, M.; Gromova, I.; Knudsen, B.R. Decreased camptothecin sensitivity of the stem-cell-like fraction of Caco2 cells correlates with an altered phosphorylation pattern of topoisomerase I. PLoS ONE 2014, 9, e99628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakobsen, A.K.; Lauridsen, K.L.; Samuel, E.B.; Proszek, J.; Knudsen, B.R.; Hager, H.; Stougaard, M. Correlation between topoisomerase I and tyrosyl-DNA phosphodiesterase 1 activities in non-small cell lung cancer tissue. Exp. Mol. Pathol. 2015, 99, 56–64. [Google Scholar] [CrossRef]
- Juul, S.; Nielsen, C.J.F.; Labouriau, R.; Roy, A.; Tesauro, C.; Jensen, P.W.; Harmsen, C.; Kristoffersen, E.L.; Chiu, Y.L.; Frohlich, R.; et al. Droplet microfluidics platform for highly sensitive and quantitative detection of malaria-causing plasmodium parasites based on enzyme activity measurement. ACS Nano 2012, 6, 10676–10683. [Google Scholar] [CrossRef] [Green Version]
- Hede, M.S.; Okorie, P.N.; Fruekilde, S.K.; Fjelstrup, S.; Thomsen, J.; Franch, O.; Tesauro, C.; Bugge, M.T.; Christiansen, M.; Picot, S.; et al. Refined method for droplet microfluidics-enabled detection of Plasmodium falciparum encoded topoisomerase i in blood from malaria patients. Micromachines 2015, 6, 1505–1513. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Champoux, J.J. Assays for the preferential binding of human topoisomerase I to supercoiled DNA. Methods Mol. Biol. 2009, 582, 49–57. [Google Scholar] [CrossRef] [Green Version]
- Alonso, C.; Fuertes, M.; Martín-Encinas, E.; Selas, A.; Rubiales, G.; Tesauro, C.; Knudssen, B.K.; Palacios, F. Novel topoisomerase I inhibitors. Syntheses and biological evaluation of phosphorus substituted quinoline derivates with antiproliferative activity. Eur. J. Med. Chem. 2018, 149, 225–237. [Google Scholar] [CrossRef]
- Lisby, M.; Olesen, J.R.; Skouboe, C.; Krogh, B.O.; Straub, T.; Boege, F.; Velmurugan, S.; Martensen, P.M.; Andersen, A.H.; Jayaram, M.; et al. Residues within the N-terminal Domain of Human Topoisomerase I Play a Direct Role in Relaxation. J. Biol. Chem. 2001, 276, 20220–20227. [Google Scholar] [CrossRef] [Green Version]
- Tesauro, C.; Juul, S.; Arno, B.; Nielsen, C.J.F.; Fiorani, P.; Frohlich, R.F.; Andersen, F.F.; Desideri, A.; Stougaard, M.; Petersen, E.; et al. Specific detection of topoisomerase i from the malaria causing P. falciparum parasite using isothermal Rolling Circle Amplification. In Proceedings of the 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August–1 September 2012; pp. 2416–2419. [Google Scholar] [CrossRef]
- Staker, B.L.; Hjerrild, K.; Feese, M.D.; Behnke, C.A.; Burgin, A.B.; Stewart, L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. USA 2002, 99, 15387–15392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pommier, Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 2006, 6, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P.; Gribaldo, S.; Gadelle, D.; Serre, M.C. Origin and evolution of DNA topoisomerases. Biochimie 2007, 89, 427–446. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. DNA relaxation and cleavage assays to study topoisomerase I inhibitors. Methods Enzymol. 2001, 340, 610–623. [Google Scholar] [CrossRef]
- Corbett, K.D.; Berger, J.M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 95–118. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petersen, K.V.; Selas, A.; Hymøller, K.M.; Mizielinski, K.; Thorsager, M.; Stougaard, M.; Alonso, C.; Palacios, F.; Pérez-Pertejo, Y.; Reguera, R.M.; et al. Simple and Fast DNA Based Sensor System for Screening of Small-Molecule Compounds Targeting Eukaryotic Topoisomerase 1. Pharmaceutics 2021, 13, 1255. https://doi.org/10.3390/pharmaceutics13081255
Petersen KV, Selas A, Hymøller KM, Mizielinski K, Thorsager M, Stougaard M, Alonso C, Palacios F, Pérez-Pertejo Y, Reguera RM, et al. Simple and Fast DNA Based Sensor System for Screening of Small-Molecule Compounds Targeting Eukaryotic Topoisomerase 1. Pharmaceutics. 2021; 13(8):1255. https://doi.org/10.3390/pharmaceutics13081255
Chicago/Turabian StylePetersen, Kamilla Vandsø, Asier Selas, Kirstine Mejlstrup Hymøller, Karol Mizielinski, Maria Thorsager, Magnus Stougaard, Concepcion Alonso, Francisco Palacios, Yolanda Pérez-Pertejo, Rosa M. Reguera, and et al. 2021. "Simple and Fast DNA Based Sensor System for Screening of Small-Molecule Compounds Targeting Eukaryotic Topoisomerase 1" Pharmaceutics 13, no. 8: 1255. https://doi.org/10.3390/pharmaceutics13081255