Multivalent Calixarene-Based Liposomes as Platforms for Gene and Drug Delivery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Liposomes
2.3. Preparation of Lipoplexes
2.4. Zeta Potential Measurements
2.5. Dynamic Light Scattering, DLS, Measurements
2.6. Agarose Gel Electrophoresis
2.7. Circular Dichroism, CD, Spectra
2.8. Atomic Force Microscopy, AFM
2.9. Electron Transmission Microscopy, TEM
2.10. In Vitro Cytotoxicity Assays
2.11. Transfection Assays
2.12. UV–Visible Spectroscopy
2.13. Encapsulation Efficiency Measurements
2.14. In Vitro Drug Release
2.15. Statistical Analysis
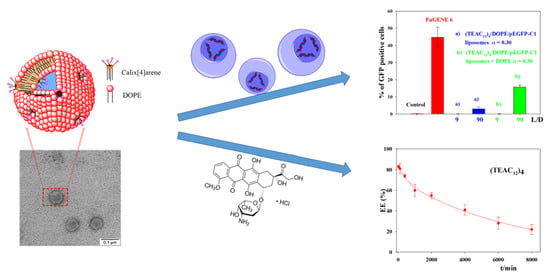
3. Results and Discussion
3.1. Calixarene-Based Liposomes
3.2. CAL/DOPE/DNA Lipoplexes
3.3. Transfection Efficiency of CAL/DOPE/pDNA Lipoplexes
3.4. Encapsulation of Doxorubicin
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, Y.P.; Jadhav, S. Novel methods for liposome preparation. Chem. Phys. Lipids 2014, 177, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.; Huang, L. Nanoparticles containing insoluble drug for cancer therapy. Biotechnol. Adv. 2014, 32, 778–788. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, B.; Gupta, R.; Gulati, M.; Singh, S.K.; Khursheed, R.; Gupta, M. The why, where, who, how, and what of the vesicular delivery systems. Adv. Colloid Interface Sci. 2019, 271, 101985. [Google Scholar] [CrossRef]
- Balazs, D.A.; Godbey, W.T. Liposomes for use in gene delivery. J. Drug Deliv. 2010, 2011, 326497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maione-Silva, L.; Gava de Castro, E.; Leite Nascimento, T.; Ramos Cintra, E.; Cleres Moreira, L.; SantanaCintra, B.A.; Campos Valadares, M.; Martins Lima, E. Ascorbic acid encapsulated into negatively charged liposomes exhibits increased skin permeation, retention and enhances collagen synthesis by fibroblasts. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bangham, A.D.; Standish, M.M.; Watkins, J.C. Diffusion of univalent ions across the lamellae of swollen phospholipids. J. Mol. Biol. 1965, 13, 238–252. [Google Scholar] [CrossRef]
- Cuomo, F.; Ceglie, S.; Miguel, M.; Lindman, B. Oral delivery of all-trans retinoic acid mediated by liposome carriers. Colloids Surf. B 2021, 201, 111655. [Google Scholar] [CrossRef]
- Logendiran, M.; Radhakrishnan, H.R.; Nagarajan, K.; Subbiah, L.; Palanisamy, S. Review on liposome based cancer theragnostics and therapy current advancements. Int. J. Pharm. Bio Sci. 2020, 11, 127–137. [Google Scholar] [CrossRef]
- Azadi, Y.; Ahmadpour, E.; Ahmadi, A. Targeting strategies in therapeutic applications of toxoplasmosis: Recent advances in liposomal vaccine delivery systems. Curr. Drug Targets 2020, 21, 541–558. [Google Scholar] [CrossRef]
- Ahmed, K.S.; Hussein, S.A.; Ali, A.H.; Korma, S.A.; Qiu, L.; Chen, J. Liposome: Composition, characterization, preparation, and recent innovation in clinical applications. J. Drug Targets 2019, 27, 742–761. [Google Scholar] [CrossRef]
- Bayat, F.; Hosseinpour-Moghadam, R.; Mehryab, F.; Fatahi, Y.; Shakeri, N.; Dinarvand, R.; Ten Hagen, T.L.M.; Haeri, A. Potential application of liposomal nanodevices for non-cancer diseases: An update on design, characterization and biopharmaceutical evaluation. Adv. Colloid Interface Sci. 2020, 277, 102121. [Google Scholar] [CrossRef]
- Daeihamed, M.; Dadashzadeh, S.; Haeri, A.; Akhlaghi, M.F. Potential of liposomes for enhancement of oral drug absorption. Curr. Drug Deliv. 2017, 14, 289–303. [Google Scholar] [CrossRef]
- Nam, J.; Beales, P.A.; Vanderlick, T.K. Giant phospholipid/block copolymer hybrid vesicles: Mixing behavior and domain formation. Langmuir 2011, 27, 1–6. [Google Scholar] [CrossRef]
- Matsumura, A.; Tsuchiye, K.; Torisoe, K.; Sakai, K.; Abe, M. Photochemical control of molecular assembly formation in a catanionic surfactant system. Langmuir 2011, 27, 1610–1617. [Google Scholar] [CrossRef]
- Devi, U.; Brown, J.R.D.; Almond, A.; Webb, S.J. Pd(II)-mediated assembly of porphyrin channels in bilayer membranes. Langmuir 2011, 7, 1448–1456. [Google Scholar] [CrossRef]
- Schuhle, D.T.; van Rijn, P.; Laurent, S.; Elst, L.V.; Muller, R.N.; Stuart, M.C.A.; Schatze, J.; Peters, J.A. Liposomes with conjugates of a calix[4]arene and a Gd-DOTA derivative on the outside surface; an efficient potential contrast agent for MRI. Chem. Commun. 2010, 46, 4399–4401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, H.-W.; Pan, Y.-C.; Guo, D.S. Assemblyenhanced molecular recognition of calix[6]arene. Supramol. Chem. 2018, 30, 562–567. [Google Scholar] [CrossRef]
- Sanabria Español, E.; Maldonado Villamil, M. Calixarenes: Generalities and their role in improving the solubility, biocompatibility, stability, bioavailability, detection, and transport of biomolecules. Biomolecules 2019, 9, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutsche, C.D. Calixarenes: An Introduction, 2nd ed.; RSC Publishing: Cambridge, UK, 2008. [Google Scholar] [CrossRef]
- Sansone, F.; Balduino, L.; Casnati, A.; Ungaro, R. Calixarenes: From biomimetic receptors to multitalented ligands for biomolecular recognition. New J. Chem. 2010, 34, 2715–2728. [Google Scholar] [CrossRef]
- Arduini, A.; Demuru, D.; Pochini, A.; Secchi, A. Recognition of quaternary ammonium cations by calix[4]arene derivatives supported on gold nanoparticles. Chem. Commun. 2005, 645–647. [Google Scholar] [CrossRef]
- Arduini, A.; Ciesa, F.; Fragassi, M.; Pochini, A.; Secchi, A. Selective synthesis of two constitutionally isomeric oriented calix[6]arene-based rotaxanes. Angew. Chem. Int. Ed. 2005, 44, 278–281. [Google Scholar] [CrossRef]
- Rudkevich, D.M. Progress in supramolecular chemistry of gases. Eur. J. Org. Chem. 2007, 2007, 3255–3270. [Google Scholar] [CrossRef]
- Rudkevich, D.M. Nanoscale molecular containers. Bull. Chem. Soc. Jpn. 2002, 75, 393–413. [Google Scholar] [CrossRef]
- Vicens, J.; Harrowfield, J. (Eds.) Calixarenes in the Nanoworld; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar] [CrossRef]
- Sansone, F.; Dudic, M.; Dpnofrio, G.; Rivetti, C.; Baldini, L.; Casnati, A.; Cellai, S.; Ungaro, R. DNA condensation and cell transfection properties of guanidinium calixarene: Dependence on macrocycle lipophilicity, size, and confrmation. J. Am. Chem. Soc. 2006, 128, 14528–14536. [Google Scholar] [CrossRef] [PubMed]
- Rodik, R.V.; Anthony, A.S.; Kalchenki, V.I.; Mely, Y.; Klymchenko, A.S. Cationic amphiphilic calixarenes to compact DNA into small nanoparticles for gene delivery. New J. Chem. 2015, 39, 1654–1664. [Google Scholar] [CrossRef]
- Ostos, F.J.; Lebrón, J.A.; López-Cornejo, P.; López-López, M.; García-Calderón, M.; García-Calderón, C.B.; Rosado, I.V.; Kalchenko, V.I.; Rodik, R.V.; Moyá, M.L. Self-aggregation in aqueous solution of amphiphilic cationic calix[4] arenes. Potential use as vectors and nanocarriers. J. Mol. Liq. 2020, 304, 112724. [Google Scholar] [CrossRef]
- Narkhede, N.; Uttam, B.; Kandi, R.; Rao, C.P. Silica-calix hybrid composite of allyl calix[4]arene linked to MCM-41 nanoparticles for sustained release of doxorubicin into cancer cells. ACS Omega 2018, 3, 229–239. [Google Scholar] [CrossRef] [Green Version]
- Ostos, F.J.; Lebrón, J.A.; Moyá, M.L.; López-López, M.; Sánchez, A.; Clavero, A.; García-Calderón, C.G.; Rosado, I.V.; López-Cornejo, P. P-Sulfocalix[6]arene as nanocarrier for controlled delivery of doxorubicin. Chem. Asian J. 2017, 12, 679–689. [Google Scholar] [CrossRef]
- Lebrón, J.A.; Ostos, F.J.; López-López, M.; Moyá, M.L.; Kardell, O.; Sánchez, A.; Carrasco, C.J.; García-Calderón, M.; García-Calderón, M.B.; Rosado, I.V.; et al. Preparation and characterization of metallomicelles of Ru(II): Cytotoxic activity and use as vectors. Colloid Surf. B 2019, 175, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Lebrón, J.A.; López-López, M.; Moyá, M.L.; Ostos, F.J.; Moreno, L.; García, D.; Moreno-Gordillo, P.; Rosado, I.V.; López-Cornejo, P. Metallo-liposomes derived from the [Ru(bpy)3]2+ complex as nanocarriers of therapeutic agents. Chemosensors 2021, 9, 90. [Google Scholar] [CrossRef]
- Barrán-Berdón, A.; Yélamos, B.; García-Río, L.; Domenech, O.; Aicart, E.; Junquera, E. Polycationc macrocyclic scaffolds as potential non-viral vectors of DNA: A multidisciplinary study. ACS Appl. Mater. Interfaces 2015, 7, 14404–14414. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, S.; Nishina, K.; Fujii, S.; Sakurai, K. The transfection efficiency of calix[4]arene-based lipids: The role of the alkyl chaim length. Biomater. Sci. 2015, 3, 317–322. [Google Scholar] [CrossRef]
- Drakalska, E.; Momekova, D.; Manolova, Y.; Budurova, D.; Momekov, G.; Genova, M.; Antonov, L.; Lambov, N.; Rangelov, S. Hybrid liposomal PEGylated calix[4]arene systems as drug delivery platforms for curcumin. Int. J. Pharm. 2014, 472, 165–174. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-J.; Chao, H.; Tan, L.-F.; Yuan, L.-X.; Ji, L.-N. Ruthenium(II) mixed-ligand complexes containing 2-(6-methyl-3-chromonyl)-imidazol[4,5-f][1,10]-phenanthroline: Synthesis, DNA-binding and photoclavage studies. Inorg. Chim. Acta 2006, 359, 3807–3814. [Google Scholar] [CrossRef]
- Secco, F.; Venturini, M.; Biver, T.; Sanchez, T.; Prado-Gotor, R.; Grueso, E. Solvent effects on the kinetics of the interaction of 1-pyrenecarboxaldehyde with calf thymus DNA. J. Phys. Chem. B 2010, 114, 4686–4691. [Google Scholar] [CrossRef]
- Asayama, S.; Nohara, A.; Negishi, Y.; Hawakami, H. Alkylimidazolium end-modified poly(ethylene glycol) to form the mono-ion complex with plasmid DNA for in vivo gene delivery. Biomacromolecules 2014, 15, 997–1001. [Google Scholar] [CrossRef]
- Zhang, H. Thin-Film hydration followed by extrusion method for liposome preparation. Methods Mol. Biol. 2017, 1522, 17–22. [Google Scholar] [CrossRef]
- Moyá, M.L.; López-López, M.; Lebrón, J.A.; Ostos, F.J.; Pérez, D.; Camacho, V.; Beck, V.; Merino-Bohórquez, V.; Camean, M.; Madinabeitia, N.; et al. Preparation and characterization of new liposomes. Bactericidal activity of cefepime encapsulated into cationic liposomes. Pharmaceutics 2019, 11, 69. [Google Scholar] [CrossRef] [Green Version]
- Meerloo, J.; Kaspers, G.J.L.; Cloos, J. Cell sensitivity assays: The MTT assay. Methods Mol. Biol. 2011, 731, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Israelachvili, J.; Mitchell, D.; Niham, B.W. Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayer. J. Chem. Soc. Faraday Trans. 2 1976, 72, 1525–1568. [Google Scholar] [CrossRef]
- Nagarajan, R.; Wang, C.-C. Theory of surfactant aggregation in water-ethylene glycol mixed solvents. Langmuir 2000, 16, 5242–5251. [Google Scholar] [CrossRef]
- Rodik, R.V.; Klymchenko, A.S.; Jain, N.; Miroshnichenko, S.I.; Richet, L.; Kalchenko, V.I.; Mely, I. Virus-sized DNA nanoparticles for gene delivery based on micelles of cationic calixarenes. Chem. Eur. J. 2011, 17, 5526–5538. [Google Scholar] [CrossRef]
- Giuliani, M.; Morbioli, I.; Sansone, F.; Casnati, A. Moulding calixarenes for biomacromolecule targeting. Chem. Commun. 2015, 51, 14140–14159. [Google Scholar] [CrossRef]
- Múñoz-Úbeda, M.; Misra, S.K.; Barrán-Berdón, A.L.; Aicart-Ramos, C.; Sierra, M.B.; Biswas, J.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. Why is less cationic lipid required to prepare lipoplexes from plasmid DNA than linear DNA in gene therapy? J. Am. Chem. Soc. 2011, 133, 18014–18017. [Google Scholar] [CrossRef]
- Ahmed, T.; Kamel, A.O.; Wettig, S.D. Interactions between DNA and Gemini surfactants: Impact on gene therapy. Part I. Nanomedicine 2016, 11, 289–306. [Google Scholar] [CrossRef]
- Kirby, A.J.; Camilleri, P.; Engberts, J.F.B.N.; Feiters, M.N.; Nolte, R.J.M.; Söderman, O.; Bergsma, M.; Bell, P.C.; Fielden, M.L.; García Rodríguez, C.I.; et al. Gemini surfactants: New synthetic vectors for gene transfection. Angew. Chem. Int. Ed. 2003, 42, 1448–1457. [Google Scholar] [CrossRef]
- Neidle, S. Nucleic Acid Structure and Recognition; Oxford University Press: New York, NY, USA, 2002; pp. 89–138. ISBN 109780198506355. [Google Scholar]
- Pietralik, Z.; Kumita, J.R.; Dobson, M.C.; Kozak, M. The influence of novel gemini surfactants containing cycloalkylside-chains on the structural phases of DNA in solution. Colloid Surf. B 2015, 131, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Bombelli, S.; Borocci, M.; Diociaiuti, F.; Faggioli, L.; Galantini, P.; Luciani, P.; Mancini, G.; Sacco, M.G. Role of the spacer of cationic gemini amphiphiles in the condensation of DNA. Langmuir 2005, 21, 10271–10274. [Google Scholar] [CrossRef]
- Bombelli, C.; Faggioli, F.; Luciani, P.; Mancini, G.; Sacco, M.G. Efficient transfection of DNA by liposomes formulated with cationic gemini amphiphiles. J. Med. Chem. 2005, 48, 5378–5382. [Google Scholar] [CrossRef]
- Marty, R.; N’soukpoe-Kossi, C.N.; Charbonneau, D.; Weinert, C.M.; Kreplack, L.; Tajmir-Riahi, H.-A. Conformational analysis of DNA complexation with cationic lipids. Nucleic Acids Res. 2009, 37, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Chaires, J.B. A thermodynamic signature for drug-DNA binding mode. Arch. Biochem. Biophys. 2006, 453, 26–31. [Google Scholar] [CrossRef]
- Yan, L.; Iwasaki, H. Thermal denaturation of plasmid DNA observed by atomic force microscopy. Jpn. J. Appl. Phys. 2002, 41, 7556–7559. [Google Scholar] [CrossRef]
- Fisicaro, E.; Compari, C.; Bacciottini, F.; Contardi, L.; Pongiluppi, E.; Barbero, N.; Viscardi, G.; Quagliotto, P.; Donofrio, G.; Kraftt, M.P. Non-viral gene delivery by highly florinated gemini bispyridinium surfactants-based DNA nanoparticles. J. Colloid Interface Sci. 2017, 102, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yang, J.; Huang, L.; Liu, D. Effect of non-ionic surfactants on the formation of DNA/emulsion complexes and emulsion-mediated gene transfer. Pharm. Res. 1996, 13, 1642–1646. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Liu, F.; Liu, D.; Huang, L. Emulsion formulations as a vector for gene delivery in vitro and vivo. Adv. Drug Deliv. Rev. 1997, 24, 265–271. [Google Scholar] [CrossRef]
- Chen, Y.; Lu, Y.; Tao, C.; Huang, J.; Zhang, H.; Yu, Y.; Zou, H.; Gao, J.; Zhong, Y. Complexes containing cationic and anionic pH-sensitive liposomes: Comparative study of factors influencing plasmid DNA gene delivery to tumors. Int. J. Nanomed. 2013, 8, 1573–1593. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, C.; Santos, R.; Cardoso, S.; Correia, S.; Oliveira, P.; Santos, M.; Moreira, P. Doxorubicin: The good, the bad and the ugly effect. Curr. Med. Chem. 2009, 16, 3267–3285. [Google Scholar] [CrossRef]
- Harada, M.; Bobe, I.; Saito, I.; Shibata, N.; Tanaka, R.; Hayashi, T.; Kato, Y. Improved anti-tumor activity of stabilized anthracycline polymeric micelle formulation, NC-6300. Cancer Sci. 2011, 102, 192–199. [Google Scholar] [CrossRef]
- Weber, P.; Wagner, M.; Schneckenburger, M. Cholesterol dependent uptake and interaction of doxorubicin MCF-7 breast cancer cells. Int. J. Mol. Sci. 2013, 14, 8358. [Google Scholar] [CrossRef]
- Walsby, E.J.; Coles, S.J.; Knapper, S.; Burnett, A.K. The topoisomerase II inhibitor bórdelo in causes cell cycle arrest and apoptosis in myeloid leukemia cells and act in synergy with cytarabine. Haematologica 2011, 96, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Thirumaran, R.; Prendergast, G.C.; Gilman, P.B. Cytotoxic chemotherapy in clinical treatment of cancer. In Cancer Immunotherapy; Prendergast, G.C., Jaffee, E.M., Eds.; Academic Press: Chicago, IL, USA, 2007; Chapter 7; pp. 101–116. [Google Scholar]
- Lefrak, E.A.; Pitha, J.; Rosenheim, S.; Gottlieb, J.A. A clinical pathological analysis of adryamicin cardiotoxycity. Cancer 1973, 32, 302–314. [Google Scholar] [CrossRef]
- Sinha, B.K.; Mimnaugh, E.G. Free radicals and anticancer drug resistance: Oxygen free radicals in the mechanisms of drug cytotoxicity and resistance by certain tumors. Free Radic. Biol. Med. 1990, 8, 567–581. [Google Scholar] [CrossRef]
- Storm, G.; van Bloois, L.; Steerenberg, P.A.; van Etten, E.; de Groot, G.; Crommelin, D.J.A. Liposome encapsulation of doxorubicin: Pharmaceutical and therapeutic aspects. J. Control. Release 1989, 9, 215–229. [Google Scholar] [CrossRef]
- Gokhale, P.C.; Radhakrishnan, B.; Husain, S.R.; Abernethy, D.R.; Sacher, R.; Dritschilo, A.; Rahman, A. An improved method of encapsulation of doxorubicin in liposomes: Pharmacological, toxicological and therapeutic evaluation. Br. J. Cancer 1996, 74, 43–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowens, L.W.; Creaven, P.J.; Greco, W.R.; Brenner, D.E.; Tung, Y.; Petrelli, N. Initial clinical (Phase I) trial of TLC-99 (Doxorubicin encapsulated liposomes). Cancer Res. 1993, 53, 2796–2802. [Google Scholar] [PubMed]
- Barenholz, Y.C. Doxil—The first FDA-approved nanodrug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef]
- Patil, R.R.; Guhagarkar, S.A.; Devarajan, P.V. Engineered nanocarriers of doxorubicin: A current update. Crit. Rev. Ther. Drug Carr. Syst. 2008, 25, 1–61. [Google Scholar] [CrossRef]
- Norouzi, M.; Yathindranath, V.; Thliveris, J.A.; Kopec, B.M.; Siahaan, T.J.; Miller, D.W. Doxorubicin-loaded iron oxide nanoparticles for glioblastoma therapy: A combinational approach for enhanced delivery of nanoparticles. Sci. Rep. 2020, 10, 11292. [Google Scholar] [CrossRef]
- Chao, Y.; Liang, Y.; Fang, G.; He, H.; Yao, Q.; Xu, H.; Chen, Y.; Tang, X. Biodegradable polymersomes as nanocarriers for doxorubicin hydrochloride: Enhanced cytotoxicity in MCF-7/ADR cells and prolonged blood circulation. Pharm. Res. 2017, 34, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Sagnella, S.M.; Duong, H.; NacMillan, A.; Boyer, C.; Whan, R.; Carroll, J.A.; Davis, T.P.; Kavallaris, M. Dextean-based doxorubicin nanocarriers with improved tumor penetration. Biomacromolecules 2014, 15, 262–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]















| α | (TEAC12)4 | (Im12)4 | (Im3Im16)2 | (Im16)4 |
|---|---|---|---|---|
| 0.1 | - | - | 1:9 | 1:9 |
| 0.2 | 1:4 | 1:4 | 1:4 | 1:4 |
| 0.3 | 1:2.33 | 1:2.33 | 1:2.33 | 1:2.33 |
| 0.4 | 1:5 | 1:5 | 1:5 | - |
| 0.45 | 1:1.22 | 1:1.22 | - | - |
| 0.5 | 1:1 | 1:1 | 1:1 | - |
| (L/D)Φ | ||||||||
|---|---|---|---|---|---|---|---|---|
| α | (TEAC12)4 | (Im12)4 | (Im16Im3)2 | (Im16)4 | ||||
| 0.10 | - | - | 6.6 a | 6.6 b | 6.9 a | 6.9 b | ||
| 0.20 | 3.8 a | 3.6 b | 3.7 a | 3.6 b | 3.6 a | 3.7 b | 3.9 a | 3.9 b |
| 0.30 | 2.8 a | 2.6 b | 2.7 a | 2.6 b | 2.6 a | 2.6 b | 2.9 a | 2.9 b |
| 0.40 | 2.3 a | 2.2 b | 2.2 a | 2.0 b | 2.1 a | 2.1 b | - | - |
| 0.45 | 2.1 a | 1.8 b | 2.0 a | 1.8 b | - | - | - | - |
| 0.50 | 2.0 a | 1.8 b | 1.9 a | 1.7 b | 1.8 a | 1.8 b | - | - |
| Hydrodynamic Diameter (DLS)/nm | Diameter (TEM)/nm | |||
|---|---|---|---|---|
| (TEAC12)4 | (Im12)4 | (TEAC12)4 | (Im12)4 | |
| Liposomes | 147 ± 4 | 157 ± 4 | 130 ± 32 | 140 ± 40 |
| Lipoplexes (L/D = 5) | 183 ± 9 | 276 ± 8 | 183 ± 39 | 223 ± 47 |
| α | EE% | LC% |
|---|---|---|
| 0.2 | 72 ± 5 | 1.85 ± 0.18 |
| 0.4 | 83 ± 1 | 4.21 ± 0.09 |
| α | dH/nm a | PDI a | dH/nm b | PDI b |
|---|---|---|---|---|
| 0.2 | 200 ± 7 | 0.23 ± 0.02 | 186 ± 6 | 0.18 ± 0.02 |
| 0.4 | 131 ± 6 | 0.163 ± 0.012 | 134 ± 6 | 0.22 ± 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebrón, J.A.; López-López, M.; García-Calderón, C.B.; V. Rosado, I.; Balestra, F.R.; Huertas, P.; Rodik, R.V.; Kalchenko, V.I.; Bernal, E.; Moyá, M.L.; et al. Multivalent Calixarene-Based Liposomes as Platforms for Gene and Drug Delivery. Pharmaceutics 2021, 13, 1250. https://doi.org/10.3390/pharmaceutics13081250
Lebrón JA, López-López M, García-Calderón CB, V. Rosado I, Balestra FR, Huertas P, Rodik RV, Kalchenko VI, Bernal E, Moyá ML, et al. Multivalent Calixarene-Based Liposomes as Platforms for Gene and Drug Delivery. Pharmaceutics. 2021; 13(8):1250. https://doi.org/10.3390/pharmaceutics13081250
Chicago/Turabian StyleLebrón, José Antonio, Manuel López-López, Clara B. García-Calderón, Ivan V. Rosado, Fernando R. Balestra, Pablo Huertas, Roman V. Rodik, Vitaly I. Kalchenko, Eva Bernal, María Luisa Moyá, and et al. 2021. "Multivalent Calixarene-Based Liposomes as Platforms for Gene and Drug Delivery" Pharmaceutics 13, no. 8: 1250. https://doi.org/10.3390/pharmaceutics13081250