Formulation and Efficacy of Catalase-Loaded Nanoparticles for the Treatment of Neonatal Hypoxic-Ischemic Encephalopathy
Abstract
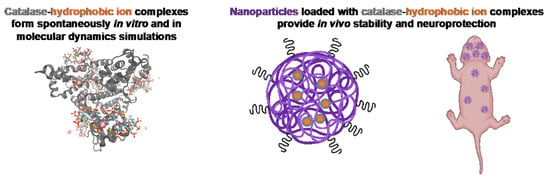
:1. Introduction
2. Materials and Methods
2.1. Preparation of HIP Complexes
2.2. Characterization of Catalase Binding Efficiency and Mass by BCA Assay
2.3. Catalase Activity Assay
2.4. Atomistic BSA/Ion Pairing (IP) Agent MD Simulations
2.5. Fraction of Surface Residues with >95% Occupancy
2.6. Nanoparticle Formulation
2.7. Nanoparticle Characterization
2.8. Animal Experiments and Ethics Statement
2.9. Vannucci Model of Unilateral HI Injury in Neonatal Rats and Drug Administration
2.10. Gross Injury Scoring and Area Loss
2.11. Immunofluorescence and Confocal Imaging
2.12. Statistical Analysis
3. Results
3.1. Effect of Ion-Pairing Agent, Molar Ratio, pH, and Buffer Ion on Complexation Efficiency
3.2. Molecular Scale Features of Protein-Ion Complexes
3.3. Effect of Nanoparticle Formulation Method on Catalase Loading and Protection
3.4. Effect of Catalase-Loaded Nanoparticles on Brain Injury Severity in Neonatal Rats
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kurinczuk, J.J.; White-Koning, M.; Badawi, N. Epidemiology of neonatal encephalopathy and hypoxic-ischaemic encephalopathy. Early Hum. Dev. 2010, 86, 329–338. [Google Scholar] [CrossRef]
- Wu, Y.W.; Backstrand, K.H.; Zhao, S.; Fullerton, H.J.; Johnston, S.C. Declining diagnosis of birth asphyxia in California: 1991–2000. Pediatrics 2004, 114, 1584–1590. [Google Scholar] [CrossRef]
- Shankaran, S.; Laptook, A.R.; Pappas, A.; McDonald, S.A.; Das, A.; Tyson, J.E.; Poindexter, B.B.; Schibler, K.; Bell, E.F.; Heyne, R.J.; et al. Effect of Depth and Duration of Cooling on Death or Disability at Age 18 Months Among Neonates with Hypoxic-Ischemic Encephalopathy: A Randomized Clinical Trial. JAMA 2017, 318, 57–67. [Google Scholar] [CrossRef]
- McPherson, R.J.; Demers, E.J.; Juul, S.E. Safety of high-dose recombinant erythropoietin in a neonatal rat model. Neonatology 2007, 91, 36–43. [Google Scholar] [CrossRef]
- Traudt, C.M.; McPherson, R.J.; Bauer, L.A.; Richards, T.L.; Burbacher, T.M.; McAdams, R.M.; Juul, S.E. Concurrent erythropoietin and hypothermia treatment improve outcomes in a term nonhuman primate model of perinatal asphyxia. Dev. Neurosci. 2013, 35, 491–503. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; He, L.; Xu, J.; Fang, Q.; Yang, L.; Xue, Y.; Wang, X.; Tang, R. Oxygen-producing catalase-based prodrug nanoparticles overcoming resistance in hypoxia-mediated chemo-photodynamic therapy. Acta Biomater. 2020, 112, 234–249. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, J.; Liang, C.; Feng, L.; Dong, Z.; Song, X.; Song, G.; Liu, Z. Drug-induced co-assembly of albumin/catalase as smart nano-theranostics for deep intra-tumoral penetration, hypoxia relieve, and synergistic combination therapy. J. Control. Release 2017, 263, 79–89. [Google Scholar] [CrossRef]
- Zhang, Q.; Tao, H.; Lin, Y.; Hu, Y.; An, H.; Zhang, D.; Feng, S.; Hu, H.; Wang, R.; Li, X.; et al. A superoxide dismutase/catalase mimetic nanomedicine for targeted therapy of inflammatory bowel disease. Biomaterials 2016, 105, 206–221. [Google Scholar] [CrossRef] [PubMed]
- Muro, S.; Cui, X.; Gajewski, C.; Murciano, J.C.; Muzykantov, V.R.; Koval, M. Slow intracellular trafficking of catalase nanoparticles targeted to ICAM-1 protects endothelial cells from oxidative stress. Am. J. Physiol. Cell Physiol. 2003, 285, C1339–C1347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armogida, M.; Spalloni, A.; Amantea, D.; Nutini, M.; Petrelli, F.; Longone, P.; Bagetta, G.; Nistico, R.; Mercuri, N.B. The protective role of catalase against cerebral ischemia In Vitro and In Vivo. Int. J. Immunopathol. Pharmacol. 2011, 24, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cheng, E.; Brooke, S.; Chang, P.; Sapolsky, R. Over-expression of antioxidant enzymes protects cultured hippocampal and cortical neurons from necrotic insults. J. Neurochem. 2003, 87, 1527–1534. [Google Scholar] [CrossRef] [Green Version]
- Singhal, A.; Morris, V.B.; Labhasetwar, V.; Ghorpade, A. Nanoparticle-mediated catalase delivery protects human neurons from oxidative stress. Cell Death Dis. 2013, 4, e903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haney, M.J.; Zhao, Y.; Li, S.; Higginbotham, S.M.; Booth, S.L.; Han, H.Y.; Vetro, J.A.; Mosley, R.L.; Kabanov, A.V.; Gendelman, H.E.; et al. Cell-mediated transfer of catalase nanoparticles from macrophages to brain endothelial, glial and neuronal cells. Nanomedicine 2011, 6, 1215–1230. [Google Scholar] [CrossRef] [Green Version]
- Patel, T.; Zhou, J.; Piepmeier, J.M.; Saltzman, W.M. Polymeric nanoparticles for drug delivery to the central nervous system. Adv. Drug Deliv. Rev. 2012, 64, 701–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, A.; Wood, T.; Chen, C.-C.; Corry, K.; Snyder, J.M.; Juul, S.E.; Parikh, P.; Nance, E. Curcumin-loaded polymeric nanoparticles for neuroprotection in neonatal rats with hypoxic-ischemic encephalopathy. Nano Res. 2018, 11, 5670–5688. [Google Scholar] [CrossRef]
- Yu, M.; Wu, J.; Shi, J.; Farokhzad, O.C. Nanotechnology for protein delivery: Overview and perspectives. J. Control. Release 2016, 240, 24–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintanar-Guerrero, D.; Allemann, E.; Fessi, H.; Doelker, E. Applications of the ion-pair concept to hydrophilic substances with special emphasis on peptides. Pharm. Res. 1997, 14, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.D.; Manning, M.C. Hydrophobic ion pairing: Altering the solubility properties of biomolecules. Pharm. Res. 1998, 15, 188–193. [Google Scholar] [CrossRef]
- Gaudana, R.; Khurana, V.; Parenky, A.; Mitra, A.K. Encapsulation of Protein-Polysaccharide HIP Complex in Polymeric Nanoparticles. J. Drug Deliv. 2011, 2011, 458128. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Gaudana, R.; Mitra, A.K. A novel approach for antibody nanocarriers development through hydrophobic ion-pairing complexation. J. Microencapsul. 2014, 31, 542–550. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.H.; Shin, E.; Wang, H.; Nolan, J.; Low, S.; Parsons, D.; Zale, S.; Ashton, S.; Ashford, M.; Ali, M.; et al. A novel in situ hydrophobic ion paring (HIP) formulation strategy for clinical product selection of a nanoparticle drug delivery system. J. Control. Release 2016, 229, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Cui, F.; Shi, K.; Cun, D.; Wang, R. Design of high payload PLGA nanoparticles containing melittin/sodium dodecyl sulfate complex by the hydrophobic ion-pairing technique. Drug Dev. Ind. Pharm. 2009, 35, 959–968. [Google Scholar] [CrossRef] [PubMed]
- Samanta, S.; Roccatano, D. Interaction of Curcumin with PEO-PPO-PEO block copolymers: A molecular dynamics study. J. Phys. Chem. B 2013, 117, 3250–3257. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Johnston, B.F.; Mackay, S.P.; Schatzlein, A.G.; Gellert, P.; Sengupta, D.; Uchegbu, I.F. In silico modelling of drug-polymer interactions for pharmaceutical formulations. J. Royal Soc. Interface 2010, 7 (Suppl. 4), S423–S433. [Google Scholar] [CrossRef] [Green Version]
- Beers, R.F., Jr.; Sizer, I.W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 1952, 195, 133–140. [Google Scholar] [CrossRef]
- Dziubla, T.D.; Karim, A.; Muzykantov, V.R. Polymer nanocarriers protecting active enzyme cargo against proteolysis. J. Control. Release 2005, 102, 427–439. [Google Scholar] [CrossRef]
- Lindahl, E.; Abraham, M.J.; Hess, B.; Van der Spoel, D. GROMACS 2020.5 Source code. Zenodo 2021. [Google Scholar] [CrossRef]
- Martinez-Rosell, G.; Giorgino, T.; De Fabritiis, G. PlayMolecule ProteinPrepare: A Web Application for Protein Preparation for Molecular Dynamics Simulations. J. Chem. Inf. Model. 2017, 57, 1511–1516. [Google Scholar] [CrossRef]
- Aliev, A.E.; Kulke, M.; Khaneja, H.S.; Chudasama, V.; Sheppard, T.D.; Lanigan, R.M. Motional timescale predictions by molecular dynamics simulations: Case study using proline and hydroxyproline sidechain dynamics. Proteins 2014, 82, 195–215. [Google Scholar] [CrossRef] [Green Version]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; Gonzalez-Outeirino, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Every, H.A.; Jiskoot, W.; Witkamp, G.-J.; Buijs, W. Molecular structure of dextran sulphate sodium in aqueous environment. J. Mol. Struct. 2018, 1156, 320–329. [Google Scholar] [CrossRef] [Green Version]
- Bayly, C.I.; Cieplak, P.; Cornell, W.; Kollman, P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Frisch, M.J.T.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision E.01; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Hopkins, C.W.; Le Grand, S.; Walker, R.C.; Roitberg, A.E. Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning. J. Chem. Theory Comput. 2015, 11, 1864–1874. [Google Scholar] [CrossRef]
- Martinez, L.; Andrade, R.; Birgin, E.G.; Martinez, J.M. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef]
- Xu, Q.; Boylan, N.J.; Cai, S.; Miao, B.; Patel, H.; Hanes, J. Scalable method to produce biodegradable nanoparticles that rapidly penetrate human mucus. J. Control. Release 2013, 170, 279–286. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.D.; Pierce, L.; Ciardiello, A.J.; Vannucci, S.J. Neonatal encephalopathy: Pre-clinical studies in neuroprotection. Biochem. Soc. Trans. 2014, 42, 564–568. [Google Scholar] [CrossRef]
- Patel, S.D.; Pierce, L.; Ciardiello, A.; Hutton, A.; Paskewitz, S.; Aronowitz, E.; Voss, H.U.; Moore, H.; Vannucci, S.J. Therapeutic hypothermia and hypoxia-ischemia in the term-equivalent neonatal rat: Characterization of a translational preclinical model. Pediatr. Res. 2015, 78, 264–271. [Google Scholar] [CrossRef]
- Wood, T.; Osredkar, D.; Puchades, M.; Maes, E.; Falck, M.; Flatebo, T.; Walloe, L.; Sabir, H.; Thoresen, M. Treatment temperature and insult severity influence the neuroprotective effects of therapeutic hypothermia. Sci. Rep. 2016, 6, 23430. [Google Scholar] [CrossRef] [Green Version]
- Wood, T.; Hobbs, C.; Falck, M.; Brun, A.C.; Loberg, E.M.; Thoresen, M. Rectal temperature in the first five hours after hypoxia-ischemia critically affects neuropathological outcomes in neonatal rats. Pediatr. Res. 2018, 83, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Kellert, B.A.; McPherson, R.J.; Juul, S.E. A comparison of high-dose recombinant erythropoietin treatment regimens in brain-injured neonatal rats. Pediatr. Res. 2007, 61, 451–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juul, S.E.; Beyer, R.P.; Bammler, T.K.; McPherson, R.J.; Wilkerson, J.; Farin, F.M. Microarray analysis of high-dose recombinant erythropoietin treatment of unilateral brain injury in neonatal mouse hippocampus. Pediatr. Res. 2009, 65, 485–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baler, K.; Martin, O.A.; Carignano, M.A.; Ameer, G.A.; Vila, J.A.; Szleifer, I. Electrostatic unfolding and interactions of albumin driven by pH changes: A molecular dynamics study. J. Phys. Chem. B 2014, 118, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Hegg, P.O. Precipitation of egg white proteins below their isoelectric points by sodium dodecyl sulphate and temperature. Biochim. Biophys. Acta 1979, 579, 73–87. [Google Scholar] [CrossRef]
- Matsuura, J.; Powers, M.E.; Manning, M.C.; Shefter, E. Structure and stability of insulin dissolved in 1-octanol. J. Am. Chem. Soc. 1993, 115, 1261–1264. [Google Scholar] [CrossRef]
- Yoo, H.S.; Choi, H.-K.; Park, T.G. Protein–fatty acid complex for enhanced loading and stability within biodegradable nanoparticles. J. Pharm. Sci. 2001, 90, 194–201. [Google Scholar] [CrossRef]
- Stigter, D.; Dill, K.A. Charge effects on folded and unfolded proteins. Biochemistry 1990, 29, 1262–1271. [Google Scholar] [CrossRef] [PubMed]
- Bartoszek, M.; Sułkowski, W. The Study of pH Influence on Bovine Liver Catalase by Means of UV-VIS Spectroscopyand Spin Labelling Method. Pol. J. Environ. Stud. 2006, 15, 41–43. [Google Scholar]
- Dai, W.G.; Dong, L.C. Characterization of physiochemical and biological properties of an insulin/lauryl sulfate complex formed by hydrophobic ion pairing. Int. J. Pharm. 2007, 336, 58–66. [Google Scholar] [CrossRef]
- Ristroph, K.D.; Prud’homme, R.K. Hydrophobic ion pairing: Encapsulating small molecules, peptides, and proteins into nanocarriers. Nanoscale Adv. 2019, 1, 4207–4237. [Google Scholar] [CrossRef]
- Yang, S.; Yuan, W.; Jin, T. Formulating protein therapeutics into particulate forms. Expert Opin. Drug Deliv. 2009, 6, 1123–1133. [Google Scholar] [CrossRef]
- Liao, R.; Pon, J.; Chungyoun, M.; Nance, E. Enzymatic protection and biocompatibility screening of enzyme-loaded polymeric nanoparticles for neurotherapeutic applications. Biomaterials 2020, 257, 120238. [Google Scholar] [CrossRef]
- Petro, M.; Jaffer, H.; Yang, J.; Kabu, S.; Morris, V.B.; Labhasetwar, V. Tissue plasminogen activator followed by antioxidant-loaded nanoparticle delivery promotes activation/mobilization of progenitor cells in infarcted rat brain. Biomaterials 2016, 81, 169–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Ling, C.L.; Pang, L.; Wang, Q.; Liu, J.X.; Wang, B.S.; Liang, J.M.; Guo, Y.Z.; Qin, J.; Wang, J.X. Direct Macromolecular Drug Delivery to Cerebral Ischemia Area using Neutrophil-Mediated Nanoparticles. Theranostics 2017, 7, 3260–3275. [Google Scholar] [CrossRef]
- Lutton, E.M.; Razmpour, R.; Andrews, A.M.; Cannella, L.A.; Son, Y.J.; Shuvaev, V.V.; Muzykantov, V.R.; Ramirez, S.H. Acute administration of catalase targeted to ICAM-1 attenuates neuropathology in experimental traumatic brain injury. Sci. Rep. 2017, 7, 3846. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, F.F.; Ferriero, D.M. Neuroprotection in the newborn infant. Clin. Perinatol. 2009, 36, 859–880. [Google Scholar] [CrossRef] [Green Version]
- Brekke, E.M.; Morken, T.S.; Wideroe, M.; Haberg, A.K.; Brubakk, A.M.; Sonnewald, U. The pentose phosphate pathway and pyruvate carboxylation after neonatal hypoxic-ischemic brain injury. J. Cereb. Blood Flow Metab. 2014, 34, 724–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishna, S.; Hutton, A.; Aronowitz, E.; Moore, H.; Vannucci, S.J. The effects of adding prophylactic phenobarbital to therapeutic hypothermia in the term-equivalent hypoxic-ischemic rat. Pediatr. Res. 2018, 83, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Fanjul, J.; Duran Fernandez-Feijoo, C.; Lopez-Abad, M.; Lopez Ramos, M.G.; Balada Caballe, R.; Alcantara-Horillo, S.; Camprubi Camprubi, M. Neuroprotection with hypothermia and allopurinol in an animal model of hypoxic-ischemic injury: Is it a gender question? PLoS ONE 2017, 12, e0184643. [Google Scholar] [CrossRef] [Green Version]
- Matchett, G.A.; Fathali, N.; Hasegawa, Y.; Jadhav, V.; Ostrowski, R.P.; Martin, R.D.; Dorotta, I.R.; Sun, X.; Zhang, J.H. Hydrogen gas is ineffective in moderate and severe neonatal hypoxia-ischemia rat models. Brain Res. 2009, 1259, 90–97. [Google Scholar] [CrossRef]
- Smith, A.L.; Rosenkrantz, T.S.; Fitch, R.H. Effects of Sex and Mild Intrainsult Hypothermia on Neuropathology and Neural Reorganization following Neonatal Hypoxic Ischemic Brain Injury in Rats. Neural Plast. 2016, 2016, 2585230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, T.R.; Gundersen, J.K.; Falck, M.; Maes, E.; Osredkar, D.; Loberg, E.M.; Sabir, H.; Walloe, L.; Thoresen, M. Variability and sex-dependence of hypothermic neuroprotection in a rat model of neonatal hypoxic-ischaemic brain injury: A single laboratory meta-analysis. Sci. Rep. 2020, 10, 10833. [Google Scholar] [CrossRef]
- Du, L.; Bayir, H.; Lai, Y.; Zhang, X.; Kochanek, P.M.; Watkins, S.C.; Graham, S.H.; Clark, R.S. Innate gender-based proclivity in response to cytotoxicity and programmed cell death pathway. J. Biol. Chem. 2004, 279, 38563–38570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demarest, T.G.; McCarthy, M.M. Sex differences in mitochondrial (dys)function: Implications for neuroprotection. J. Bioenerg. Biomembr. 2015, 47, 173–188. [Google Scholar] [CrossRef]





| Protein | Formulation Method | Number Mean ± SEM (nm) | PDI | ζ-Potential ± SEM (mV) |
|---|---|---|---|---|
| Catalase | Nanoprecipitation | 115.8 ± 1.9 | 0.17 | −2.3 ± 0.2 |
| Catalase | S/O/W emulsion | 125.4 ± 5.2 | 0.25 | −5.6 ± 0.4 |
| Bovine serum albumin | Nanoprecipitation | 106.5 ± 5.4 | 0.13 | −2.6 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joseph, A.; Nyambura, C.W.; Bondurant, D.; Corry, K.; Beebout, D.; Wood, T.R.; Pfaendtner, J.; Nance, E. Formulation and Efficacy of Catalase-Loaded Nanoparticles for the Treatment of Neonatal Hypoxic-Ischemic Encephalopathy. Pharmaceutics 2021, 13, 1131. https://doi.org/10.3390/pharmaceutics13081131
Joseph A, Nyambura CW, Bondurant D, Corry K, Beebout D, Wood TR, Pfaendtner J, Nance E. Formulation and Efficacy of Catalase-Loaded Nanoparticles for the Treatment of Neonatal Hypoxic-Ischemic Encephalopathy. Pharmaceutics. 2021; 13(8):1131. https://doi.org/10.3390/pharmaceutics13081131
Chicago/Turabian StyleJoseph, Andrea, Chris W. Nyambura, Danielle Bondurant, Kylie Corry, Denise Beebout, Thomas R. Wood, Jim Pfaendtner, and Elizabeth Nance. 2021. "Formulation and Efficacy of Catalase-Loaded Nanoparticles for the Treatment of Neonatal Hypoxic-Ischemic Encephalopathy" Pharmaceutics 13, no. 8: 1131. https://doi.org/10.3390/pharmaceutics13081131