Development of siRNA-Loaded Lipid Nanoparticles Targeting Long Non-Coding RNA LINC01257 as a Novel and Safe Therapeutic Approach for t(8;21) Pediatric Acute Myeloid Leukemia
Abstract
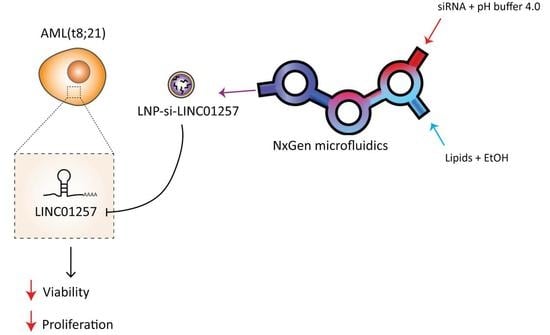
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Differential Expression and Survival Analysis
2.3. ChIP-seq Data
2.4. Cell Culture
2.5. AML Patient Samples
2.6. Cell Fractionation
2.7. RNA Isolation and qPCR
2.8. siRNA Electroporation, Cell Counts and Apoptosis Assay
2.9. Generation of LNP-siRNA
2.10. Cryogenic Electron Microscopy Imaging
2.11. Fluorescence Microscopy Analysis of LNP-siRNA Uptake in AML Cells
2.12. LNP-siRNA Association with Cells
2.13. LNP-siRNA Treatment of AML Cells and PBMCs
2.14. Statistical Analysis
3. Results
3.1. Identification of lncRNAs Specific to Pediatric t(8;21) AML
3.2. LINC01257 Knockdown Impairs AML Proliferation and Reduces Cell Viability In Vitro
3.3. LINC01257 Is Specifically Expressed in t(8;21) AML and Absent in Healthy Cells
3.4. Anti-LINC01257 siRNA-Loaded LNPs Are Efficiently Taken Up by Kasumi-1 Cells In Vitro
3.5. si-LINC01257 siRNA-Loaded LNPs Efficiently Ablate LINC01257 Expression
3.6. LNP-si-LINC01257 Impairs Kasumi-1 Cell Proliferation without Affecting Healthy PBMCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Morais, R.V.; de Souza, M.V.; Silva, K.A.D.S.; Santiago, P.; Lorenzoni, M.C.; Lorea, C.F.; Junior, C.G.D.C.; Taniguchi, A.N.R.; Scherer, F.F.; Michalowski, M.B.; et al. Epidemiological evaluation and survival of children with acute myeloid leukemia. J. Pediatr. 2021, 97, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Reilly, A.; Kersun, L.S.; Prak, E.L.; Boyer, J.; McDonald, K.; Jawad, A.F.; Sullivan, K. Immunologic Consequences of Chemotherapy for Acute Myeloid Leukemia. J. Pediatr. Hematol. 2013, 35, 46–53. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.I. How close are we to CAR T-cell therapy for AML? Best Pract. Res. Clin. Haematol. 2019, 32, 101104. [Google Scholar] [CrossRef] [PubMed]
- Kuhlen, M.; Klusmann, J.-H.; Hoell, J.I. Molecular Approaches to Treating Pediatric Leukemias. Front. Pediatr. 2019, 7, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J. Oncogenic chromosomal translocations and human cancer (Review). Oncol. Rep. 2013, 30, 2011–2019. [Google Scholar] [CrossRef] [Green Version]
- Pikman, Y.; Stegmaier, K. Targeted therapy for fusion-driven high-risk acute leukemia. Blood 2018, 132, 1241–1247. [Google Scholar] [CrossRef] [Green Version]
- Martens, J.; Stunnenberg, H.G. The molecular signature of oncofusion proteins in acute myeloid leukemia. FEBS Lett. 2010, 584, 2662–2669. [Google Scholar] [CrossRef] [Green Version]
- Tonks, A.; Pearn, L.; Musson, M.; Gilkes, A.; Mills, K.I.; Burnett, A.K.; Darley, R.L. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia. Leukemia 2007, 21, 2495–2505. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Sun, L.-Y.; Wang, W.-T.; Sun, Y.-M.; Chen, Y.-Q. Non-coding RNAs in cancers with chromosomal rearrangements: The signatures, causes, functions and implications. J. Mol. Cell Biol. 2019, 11, 886–898. [Google Scholar] [CrossRef]
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol. Med. 2018, 24, 257–277. [Google Scholar] [CrossRef]
- Delás, M.J.; Sabin, L.R.; Dolzhenko, E.; Knott, S.R.; Maravilla, E.M.; Jackson, B.T.; Wild, S.A.; Kovacevic, T.; Stork, E.M.; Zhou, M.; et al. lncRNA requirements for mouse acute myeloid leukemia and normal differentiation. eLife 2017, 6, e25607. [Google Scholar] [CrossRef]
- Zhang, X.; Goel, V.; Robbie, G.J. Pharmacokinetics of Patisiran, the First Approved RNA Interference Therapy in Patients with Hereditary Transthyretin-Mediated Amyloidosis. J. Clin. Pharmacol. 2019, 60, 573–585. [Google Scholar] [CrossRef] [Green Version]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef]
- Gao, H.; Cheng, R.; Santos, H.A. Nanoparticle-mediated siRNA delivery systems for cancer therapy. View 2021, 2, 20200111. [Google Scholar] [CrossRef]
- Jyotsana, N.; Sharma, A.; Chaturvedi, A.; Scherr, M.; Kuchenbauer, F.; Sajti, L.; Barchanski, A.; Lindner, R.; Noyan, F.; Sühs, K.-W.; et al. RNA interference efficiently targets human leukemia driven by a fusion oncogene in vivo. Leukemia 2017, 32, 224–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitazoe, K.; Wang, J.; Kaji, N.; Okamoto, Y.; Tokeshi, M.; Kogure, K.; Harashima, H.; Baba, Y. A touch-and-go lipid wrapping technique in microfluidic channels for rapid fabrication of multifunctional envelope-type gene delivery nanodevices. Lab Chip 2011, 11, 3256–3262. [Google Scholar] [CrossRef] [PubMed]
- Maeki, M.; Kimura, N.; Sato, Y.; Harashima, H.; Tokeshi, M. Advances in microfluidics for lipid nanoparticles and extracellular vesicles and applications in drug delivery systems. Adv. Drug Deliv. Rev. 2018, 128, 84–100. [Google Scholar] [CrossRef]
- Jyotsana, N.; Sharma, A.; Chaturvedi, A.; Budida, R.; Scherr, M.; Kuchenbauer, F.; Lindner, R.; Noyan, F.; Sühs, K.-W.; Stangel, M.; et al. Lipid nanoparticle-mediated siRNA delivery for safe targeting of human CML in vivo. Ann. Hematol. 2019, 98, 1905–1918. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Therneau, T. A Package for Survival Analysis in R. R package version 3.2-13. 2021. Available online: https://CRAN.R-project.org/package=survival (accessed on 13 October 2021).
- Hazan-Halevy, I.; Rosenblum, D.; Ramishetti, S.; Peer, D. Systemic Modulation of Lymphocyte Subsets Using siRNAs Delivered via Targeted Lipid Nanoparticles. Methods Mol. Biol. 2019, 1974, 151–159. [Google Scholar]
- Schuback, H.L.; Arceci, R.J.; Meshinchi, S. Somatic Characterization of Pediatric Acute Myeloid Leukemia Using Next-Generation Sequencing. Semin. Hematol. 2013, 50, 325–332. [Google Scholar] [CrossRef]
- Zuo, X.; Chen, Z.; Gao, W.; Zhang, Y.; Wang, J.; Wang, J.; Cao, M.; Cai, J.; Wu, J.; Wang, X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020, 13, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhong, Y.; Yang, B.; Zhu, Y.; Zhu, X.; Xia, Z.; Xu, J.; Xu, L. LINC00958 facilitates cervical cancer cell proliferation and metastasis by sponging miR-625-5p to upregulate LRRC8E expression. J. Cell. Biochem. 2020, 121, 2500–2509. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Xie, M.; Zhang, Z. LINC00958 Involves in Bladder Cancer Through Sponging miR-378a-3p to Elevate IGF1R. Cancer Biother. Radiopharm. 2020, 35, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Tyner, J.W. RNAi Screening of Leukemia Cells Using Electroporation. Methods Mol. Biol. 2016, 1470, 85–94. [Google Scholar] [PubMed] [Green Version]
- Lonetti, A.; Pession, A.; Masetti, R. Targeted Therapies for Pediatric AML: Gaps and Perspective. Front. Pediatr. 2019, 7, 463. [Google Scholar] [CrossRef] [PubMed]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef]
- Gutschner, T.; Hämmerle, M.; Eißmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Groß, M.; et al. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef] [Green Version]
- Vaidya, A.M.; Sun, Z.; Ayat, N.; Schilb, A.; Liu, X.; Jiang, H.; Sun, D.; Scheidt, J.; Qian, V.; He, S.; et al. Systemic Delivery of Tumor-Targeting siRNA Nanoparticles against an Oncogenic LncRNA Facilitates Effective Triple-Negative Breast Cancer Therapy. Bioconjugate Chem. 2019, 30, 907–919. [Google Scholar] [CrossRef]
- Schoch, C.; Kohlmann, A.; Schnittger, S.; Brors, B.; Dugas, M.; Mergenthaler, S.; Kern, W.; Hiddemann, W.; Eils, R.; Haferlach, T. Acute myeloid leukemias with reciprocal rearrangements can be distinguished by specific gene expression profiles. Proc. Natl. Acad. Sci. USA 2002, 99, 10008–10013. [Google Scholar] [CrossRef] [Green Version]
- Bullinger, L.; Döhner, K.; Bair, E.; Fröhling, S.; Schlenk, R.F.; Tibshirani, R.; Döhner, H.; Pollack, J.R. Use of Gene-Expression Profiling to Identify Prognostic Subclasses in Adult Acute Myeloid Leukemia. N. Engl. J. Med. 2004, 350, 1605–1616. [Google Scholar] [CrossRef] [Green Version]
- Virtaneva, K.; Wright, F.A.; Tanner, S.M.; Yuan, B.; Lemon, W.J.; Caligiuri, M.A.; Bloomfield, C.D.; de la Chapelle, A.; Krahe, R. Expression profiling reveals fundamental biological differences in acute myeloid leukemia with isolated trisomy 8 and normal cytogenetics. Proc. Natl. Acad. Sci. USA 2001, 98, 1124–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connerty, P.; Lock, R.; De Bock, C.E. Long Non-coding RNAs: Major Regulators of Cell Stress in Cancer. Front. Oncol. 2020, 10, 285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, M.-C.; Ni, J.-J.; Cui, W.-Y.; Wang, B.-Y.; Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar]
- Dizaji, B.F. Strategies to target long non-coding RNAs in cancer treatment: Progress and challenges. Egypt. J. Med. Hum. Genet. 2020, 21, 1–15. [Google Scholar] [CrossRef]
- Tatiparti, K.; Sau, S.; Kashaw, S.K.; Iyer, A.K. siRNA Delivery Strategies: A Comprehensive Review of Recent Developments. Nanomaterials 2017, 7, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotelianski, V.; Zatsepin, T.S.; Kotelevtsev, Y. Lipid nanoparticles for targeted siRNA delivery—Going from bench to bedside. Int. J. Nanomed. 2016, 11, 3077–3086. [Google Scholar] [CrossRef] [Green Version]
- Weinstein, S.; Toker, I.A.; Emmanuel, R.; Ramishetti, S.; Hazan-Halevy, I.; Rosenblum, D.; Goldsmith, M.; Abraham, A.; Benjamini, O.; Bairey, O.; et al. Harnessing RNAi-based nanomedicines for therapeutic gene silencing in B-cell malignancies. Proc. Natl. Acad. Sci. USA 2016, 113, E16–E22. [Google Scholar] [CrossRef] [Green Version]
- Kedmi, R.; Veiga, N.; Ramishetti, S.; Goldsmith, M.; Rosenblum, D.; Dammes, N.; Hazan-Halevy, I.; Nahary, L.; Leviatan-Ben-Arye, S.; Harlev, M.; et al. A modular platform for targeted RNAi therapeutics. Nat. Nanotechnol. 2018, 13, 214–219. [Google Scholar] [CrossRef]
- Yonezawa, S.; Koide, H.; Asai, T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv. Drug Deliv. Rev. 2020, 154–155, 64–78. [Google Scholar] [CrossRef]
- Belliveau, N.; Huft, J.; Lin, P.J.; Chen, S.; Leung, A.K.; Leaver, T.J.; Wild, A.W.; Lee, J.B.; Taylor, R.J.; Tam, Y.K.; et al. Microfluidic Synthesis of Highly Potent Limit-size Lipid Nanoparticles for In Vivo Delivery of siRNA. Mol. Ther.—Nucleic Acids 2012, 1, e37. [Google Scholar] [CrossRef]
- Roces, C.B.; Lou, G.; Jain, N.; Abraham, S.; Thomas, A.; Halbert, G.W.; Perrie, Y. Manufacturing Considerations for the Development of Lipid Nanoparticles Using Microfluidics. Pharmaceutics 2020, 12, 1095. [Google Scholar] [CrossRef]
- Thomas, A.; Garg, S.M.; De Souza, R.A.G.; Ouellet, E.; Tharmarajah, G.; Reichert, D.; Ordobadi, M.; Ip, S.; Ramsay, E.C. Microfluidic Production and Application of Lipid Nanoparticles for Nucleic Acid Transfection. Methods Mol. Biol. 2018, 1792, 193–203. [Google Scholar] [PubMed]
- Lorenzer, C.; Dirin, M.; Winkler, A.-M.; Baumann, V.; Winkler, J. Going beyond the liver: Progress and challenges of targeted delivery of siRNA therapeutics. J. Control Release 2015, 203, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Z.; Al Zaki, A.; Hui, J.Z.; Muzykantov, V.R.; Tsourkas, A. Multifunctional Nanoparticles: Cost Versus Benefit of Adding Targeting and Imaging Capabilities. Sciences 2012, 338, 903–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, H.Ø.; Roug, A.S.; Nielsen, K.; Søndergaard, C.S.; Hokland, P. Nonviral transfection of leukemic primary cells and cells lines by siRNA—A direct comparison between Nucleofection and Accell delivery. Exp. Hematol. 2011, 39, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Witzigmann, D.; Kulkarni, J.A.; Leung, J.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv. Drug Deliv. Rev. 2020, 159, 344–363. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Connerty, P.; Moles, E.; de Bock, C.E.; Jayatilleke, N.; Smith, J.L.; Meshinchi, S.; Mayoh, C.; Kavallaris, M.; Lock, R.B. Development of siRNA-Loaded Lipid Nanoparticles Targeting Long Non-Coding RNA LINC01257 as a Novel and Safe Therapeutic Approach for t(8;21) Pediatric Acute Myeloid Leukemia. Pharmaceutics 2021, 13, 1681. https://doi.org/10.3390/pharmaceutics13101681
Connerty P, Moles E, de Bock CE, Jayatilleke N, Smith JL, Meshinchi S, Mayoh C, Kavallaris M, Lock RB. Development of siRNA-Loaded Lipid Nanoparticles Targeting Long Non-Coding RNA LINC01257 as a Novel and Safe Therapeutic Approach for t(8;21) Pediatric Acute Myeloid Leukemia. Pharmaceutics. 2021; 13(10):1681. https://doi.org/10.3390/pharmaceutics13101681
Chicago/Turabian StyleConnerty, Patrick, Ernest Moles, Charles E. de Bock, Nisitha Jayatilleke, Jenny L. Smith, Soheil Meshinchi, Chelsea Mayoh, Maria Kavallaris, and Richard B. Lock. 2021. "Development of siRNA-Loaded Lipid Nanoparticles Targeting Long Non-Coding RNA LINC01257 as a Novel and Safe Therapeutic Approach for t(8;21) Pediatric Acute Myeloid Leukemia" Pharmaceutics 13, no. 10: 1681. https://doi.org/10.3390/pharmaceutics13101681