Strengths and Challenges of Secretory Ribonucleases as AntiTumor Agents
Abstract
:1. Introduction
2. Secretory Ribonucleases Display a Vast Array of Functions
3. Mechanisms of Antitumor Action of Secretory RNases
4. Regulatory RNAs are Key Targets for Different Antitumor RNases
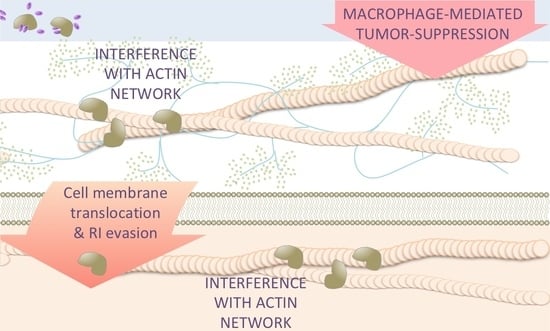
5. Effects of Antitumor RNases on Target Cells beyond RNA Degradation
6. Antitumor RNases Exert Pleiotropic Effects on Cancer Cells
7. Natural and Modified RNases as Antitumor Drugs: Concerns and Opportunities
7.1. Nanocarriers and Nanostructures to Strengthen the Efficiency of Antitumor RNases
7.2. Modification of RNases to Increase Their Pharmacokinetics and Antitumor Potency
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Si, X.; Ma, S.; Xu, Y.; Zhang, D.; Shen, N.; Yu, H.; Zhang, Y.; Song, W.; Tang, Z.; Zhu, X. Hypoxia-sensitive supramolecular nanogels for the cytosolic delivery of ribonuclease A as a breast cancer therapeutic. J. Control. Release 2020, 320, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Fan, Q.; Wang, H.; Cheng, Y. Polymers for cytosolic protein delivery. Biomaterials 2019, 218, 119358. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wan, T.; Wang, H.; Zhang, S.; Ping, Y.; Cheng, Y. A boronic acid–rich dendrimer with robust and unprecedented efficiency for cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci. Adv. 2019, 5, eaaw8922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leader, B.; Baca, Q.J.; Golan, D.E. Protein therapeutics: A summary and pharmacological classification. Nat. Rev. Drug Discov. 2008, 7, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Gotte, G.; Menegazzi, M. Biological Activities of Secretory RNases: Focus on Their Oligomerization to Design Antitumor Drugs. Front. Immunol. 2019, 10, 2626. [Google Scholar] [CrossRef] [Green Version]
- Luhtala, N.; Parker, R. T2 Family ribonucleases: Ancient enzymes with diverse roles. Trends Biochem. Sci. 2010, 35, 253–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löffler, A.; Abel, S.; Jost, W.; Beintema, J.J.; Glund, K. Phosphate-Regulated Induction of Intracellular Ribonucleases in Cultured Tomato (Lycopersicon esculentum) Cells. Plant Physiol. 1992, 98, 1472–1478. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.K.; Paz, E.; Kutsher, Y.; Reuveni, M.; Lers, A. Tomato T2 ribonuclease LE is involved in the response to pathogens. Mol. Plant Pathol. 2020, 21, 895–906. [Google Scholar] [CrossRef]
- Hillwig, M.S.; Contento, A.L.; Meyer, A.; Ebany, D.; Bassham, D.C.; MacIntosh, G.C. RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc. Natl. Acad. Sci. USA 2011, 108, 1093–1098. [Google Scholar] [CrossRef] [Green Version]
- Megel, C.; Hummel, G.; Lalande, S.; Ubrig, E.; Cognat, V.; Morelle, G.; Salinas-Giegé, T.; Duchêne, A.-M.; Maréchal-Drouard, L. Plant RNases T2, but not Dicer-like proteins, are major players of tRNA-derived fragments biogenesis. Nucleic Acids Res. 2019, 47, 941–952. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Lee, H.S.; Karunanandaa, B.; Kao, T.H. Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell 1994, 6, 1021–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClure, B.; Cruz-García, F.; Romero, C. Compatibility and incompatibility in S-RNase-based systems. Ann. Bot. 2011, 108, 647–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacIntosh, G.C.; Castandet, B. Organellar and Secretory Ribonucleases: Major Players in Plant RNA Homeostasis. Plant Physiol. 2020, 183, 1438–1452. [Google Scholar] [CrossRef] [PubMed]
- MacIntosh, G.C.; Bariola, P.A.; Newbigin, E.; Green, P.J. Characterization of Rny1, the Saccharomyces cerevisiae member of the T2 RNase family of RNases: Unexpected functions for ancient enzymes? Proc. Natl. Acad. Sci. USA 2001, 98, 1018–1023. [Google Scholar] [CrossRef] [Green Version]
- McGugan, G.C.; Joshi, M.B.; Dwyer, D.M. Identification and Biochemical Characterization of Unique Secretory Nucleases of the Human Enteric Pathogen, Entamoeba histolytica. J. Biol. Chem. 2007, 282, 31789–31802. [Google Scholar] [CrossRef] [Green Version]
- Bruschke, C.J.; Hulst, M.M.; Moormann, R.J.; Van Rijn, P.A.; Van Oirschot, J.T. Glycoprotein Erns of pestiviruses induces apoptosis in lymphocytes of several species. J. Virol. 1997, 71, 6692–6696. [Google Scholar] [CrossRef] [Green Version]
- Fontaine, B.M.; Martin, K.S.; Garcia-Rodriguez, J.M.; Jung, C.; Briggs, L.; Southwell, J.E.; Jia, X.; Weinert, E.E. RNase I regulates Escherichia coli 2′,3′-cyclic nucleotide monophosphate levels and biofilm formation. Biochem. J. 2018, 475, 1491–1506. [Google Scholar] [CrossRef]
- Fredens, K.; Dahl, R.; Venge, P. The Gordon phenomenon induced by the eosinophil cationic protein and eosinophil protein X. J. Allergy Clin. Immunol. 1982, 70, 361–366. [Google Scholar] [CrossRef]
- Durack, D.T.; Ackerman, S.J.; Loegering, D.A.; Gleich, G.J. Purification of human eosinophil-derived neurotoxin. Proc. Natl. Acad. Sci. USA 1981, 78, 5165–5169. [Google Scholar] [CrossRef] [Green Version]
- Tamburrini, M.; Scala, G.; Verde, C.; Ruocco, M.R.; Parente, A.; Venuta, S.; D’Alessio, G. Immunosuppressive activity of bovine seminal RNase on T-cell proliferation. JBIC J. Biol. Inorg. Chem. 1990, 190, 145–148. [Google Scholar] [CrossRef]
- Kim, J.S.; Soucek, J.; Matousek, J.; Raines, R.T. Catalytic activity of bovine seminal ribonuclease is essential for its immunosuppressive and other biological activities. Biochem. J. 1995, 308, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Xu, Z. Mechanisms of action of angiogenin. Acta Biochim. Biophys. Sin. 2008, 40, 619–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fett, J.W.; Strydom, D.J.; Lobb, R.R.; Alderman, E.M.; Bethune, J.L.; Riordan, J.F.; Vallee, B.L. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry 1985, 24, 5480–5486. [Google Scholar] [CrossRef] [PubMed]
- Strydom, D.J. The angiogenins. Cell. Mol. Life Sci. 1998, 54, 811–824. [Google Scholar] [CrossRef]
- Li, S.; Sheng, J.; Hu, J.K.; Yu, W.; Kishikawa, H.; Hu, M.G.; Shima, K.; Wu, D.; Xu, Z.; Xin, W.; et al. Ribonuclease 4 protects neuron degeneration by promoting angiogenesis, neurogenesis, and neuronal survival under stress. Angiogenesis 2012, 16, 387–404. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Li, J.; Moussaoui, M.; Boix, E. Immune Modulation by Human Secreted RNases at the Extracellular Space. Front. Immunol. 2018, 9, 1012. [Google Scholar] [CrossRef] [Green Version]
- Baranzini, N.; Pedrini, E.; Girardello, R.; Tettamanti, G.; De Eguileor, M.; Taramelli, R.; Grimaldi, A.; Acquati, F. Human recombinant RNASET2-induced inflammatory response and connective tissue remodeling in the medicinal leech. Cell Tissue Res. 2017, 368, 337–351. [Google Scholar] [CrossRef]
- Domachowske, J.B.; Dyer, K.D.; Bonville, C.A.; Rosenberg, H.F. Recombinant Human Eosinophil-Derived Neurotoxin/RNase 2 Functions as an Effective Antiviral Agent against Respiratory Syncytial Virus. J. Infect. Dis. 1998, 177, 1458–1464. [Google Scholar] [CrossRef] [Green Version]
- Domachowske, J.B.; Dyer, K.D.; Adams, A.G.; Leto, T.L.; Rosenberg, H.F. Eosinophil cationic protein/RNase 3 is another RNase A-family ribonuclease with direct antiviral activity. Nucleic Acids Res. 1998, 26, 3358–3363. [Google Scholar] [CrossRef]
- Rugeles, M.T.; Trubey, C.M.; Bedoya, V.I.; Pinto, L.A.; Oppenheim, J.J.; Rybak, S.M.; Shearer, G.M. Ribonuclease is partly responsible for the HIV-1 inhibitory effect activated by HLA alloantigen recognition. AIDS 2003, 17, 481–486. [Google Scholar] [CrossRef]
- Boix, E.; Torrent, M.; Sánchez, D.; Nogués, V.M. The Antipathogen Activities of Eosinophil Cationic Protein. Curr. Pharm. Biotechnol. 2008, 9, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Pulido, D.; Arranz-Trullén, J.; Prats-Ejarque, G.; Velázquez, D.; Torrent, M.; Moussaoui, M.; Boix, E. Insights into the Antimicrobial Mechanism of Action of Human RNase6: Structural Determinants for Bacterial Cell Agglutination and Membrane Permeation. Int. J. Mol. Sci. 2016, 17, 552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, L.; Arranz-Trullén, J.; Prats-Ejarque, G.; Pulido, D.; Bhakta, S.; Boix, E. Human Antimicrobial RNases Inhibit Intracellular Bacterial Growth and Induce Autophagy in Mycobacteria-Infected Macrophages. Front. Immunol. 2019, 10, 1500. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Schröder, J.-M. RNase 7, a Novel Innate Immune Defense Antimicrobial Protein of Healthy Human Skin. J. Biol. Chem. 2002, 277, 46779–46784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudolph, B.; Podschun, R.; Sahly, H.; Schubert, S.; Schröder, J.M.; Harder, J. Identification of RNase 8 as a Novel Human Antimicrobial Protein. Antimicrob. Agents Chemother. 2006, 50, 3194–3196. [Google Scholar] [CrossRef] [Green Version]
- Patutina, O.; Mирoнoва, Н.Л.; Ryabchikova, E.; Popova, N.; Nikolin, V.; Kaledin, V.; Vlassov, V.V.; Zenkova, M. Inhibition of metastasis development by daily administration of ultralow doses of RNase A and DNase I. Biochimie 2011, 93, 689–696. [Google Scholar] [CrossRef]
- Laccetti, P.; Portella, G.; Mastronicola, M.R.; Russo, A.; Piccoli, R.; D’Alessio, G.; Vecchio, G. In vivo and in vitro growth-inhibitory effect of bovine seminal ribonuclease on a system of rat thyroid epithelial transformed cells and tumors. Cancer Res. 1992, 52, 4582–4586. [Google Scholar]
- Lee, J.E.; Raines, R.T. Cytotoxicity of Bovine Seminal Ribonuclease: Monomer versus Dimer. Biochemistry 2005, 44, 15760–15767. [Google Scholar] [CrossRef]
- Ardelt, W.; Mikulski, S.M.; Shogen, K. Amino acid sequence of an anti-tumor protein from Rana pipiens oocytes and early embryos. Homology to pancreatic ribonucleases. J. Biol. Chem. 1991, 266, 245–251. [Google Scholar] [CrossRef]
- Darzynkiewicz, Z.; Carter, S.P.; Mikulski, S.M.; Ardelt, W.J.; Shogen, K. Cytostatic and Cytotoxic Effects of Pannon (P-30 Protein), A Novel Anticancer Agent. Cell Prolif. 1988, 21, 169–182. [Google Scholar] [CrossRef]
- Ardelt, W.; Shogen, K.; Darzynkiewicz, Z. Onconase and Amphinase, the Antitumor Ribonucleases from Rana pipiens Oocytes. Curr. Pharm. Biotechnol. 2008, 9, 215–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kariya, Y.; Tatsuta, T.; Sugawara, S.; Kariya, Y.; Nitta, K.; Hosono, M. RNase activity of sialic acid-binding lectin from bullfrog eggs drives antitumor effect via the activation of p38 MAPK to caspase-3/7 signaling pathway in human breast cancer cells. Int. J. Oncol. 2016, 49, 1334–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatsuta, T.; Satoh, T.; Sugawara, S.; Hara, A.; Hosono, M. Sialic acid-binding lectin from bullfrog eggs inhibits human malignant mesothelioma cell growth in vitro and in vivo. PLoS ONE 2018, 13, e0190653. [Google Scholar] [CrossRef] [Green Version]
- Acquati, F.; Possati, L.; Ferrante, L.; Campomenosi, P.; Talevi, S.; Bardelli, S.; Margiotta, C.; Russo, A.; Bortoletto, E.; Rocchetti, R.; et al. Tumor and metastasis suppression by the human RNASET2 gene. Int. J. Oncol. 2005, 26, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Roiz, L.; Smirnoff, P.; Bar-Eli, M.; Schwartz, B.; Shoseyov, O. ACTIBIND, an actin-binding fungal T2-RNase with antiangiogenic and anticarcinogenic characteristics. Cancer 2006, 106, 2295–2308. [Google Scholar] [CrossRef] [Green Version]
- Makarov, A.A.; Kolchinsky, A.; Ilinskaya, O.N. Binase and other microbial RNases as potential anticancer agents. BioEssays 2008, 30, 781–790. [Google Scholar] [CrossRef]
- Benito, A.; Ribó, M.; Vilanova, M. Molecular BioSystems; The Royal Society of Chemistry: London, UK, 11 October 2005; pp. 294–302. [Google Scholar]
- Szachowicz-Petelska, B.; Dobrzyńska, I.; Skrodzka, M.; Darewicz, B.; Figaszewski, Z.A.; Kudelski, J. Phospholipid Composition and Electric Charge in Healthy and Cancerous Parts of Human Kidneys. J. Membr. Biol. 2013, 246, 421–425. [Google Scholar] [CrossRef] [Green Version]
- Nitta, K.; Takayanagi, G.; Kawauchi, H.; Hakomori, S. Isolation and characterization of Rana catesbeiana lectin and demonstration of the lectin-binding glycoprotein of rodent and human tumor cell membranes. Cancer Res. 1987, 47, 4877–4883. [Google Scholar]
- Wu, Y.; Mikulski, S.M.; Ardelt, W.; Rybak, S.M.; Youle, R.J. A Cytotoxic Ribonuclease. Study of the Mechanism of Onconase Citotoxicity. J. Biol. Chem. 1993, 268, 10686–10693. [Google Scholar] [CrossRef]
- Haigis, M.C.; Raines, R.T. Secretory ribonucleases are internalized by a dynamin-independent endocytic pathway. J. Cell Sci. 2003, 116, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Bracale, A.; Castaldi, F.; Nitsch, L.; D’Alessio, G. A role for the intersubunit disulfides of seminal RNase in the mechanism of its antitumor action. JBIC J. Biol. Inorg. Chem. 2003, 270, 1980–1987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuentes, H.C.; Kalacheva, N.V.; Mukhametshina, R.T.; Zelenikhin, P.V.; Kolpakov, A.I.; Barreto, G.; Preissner, K.T.; Ilinskaya, O.N. Binase penetration into alveolar epithelial cells does not induce cell death. Biochem. Suppl. Ser. B Biomed. Chem. 2012, 6, 317–321. [Google Scholar] [CrossRef]
- Mironova, N.; Patutina, O.; Brenner, E.; Kurilshikov, A.; Vlassov, V.; Zenkova, M. MicroRNA Drop in the Bloodstream and MicroRNA Boost in the Tumour Caused by Treatment with Ribonuclease A Leads to an Attenuation of Tumour Malignancy. PLoS ONE 2013, 8, e83482. [Google Scholar] [CrossRef] [PubMed]
- Bracale, A.; Spalletti-Cernia, D.; Mastronicola, M.; Castaldi, F.; Mannucci, R.; Nitsch, L.; D’alessio, G. Essential stations in the intracellular pathway of cytotoxic bovine seminal ribonuclease. Biochem. J. 2002, 362, 553. [Google Scholar] [CrossRef] [PubMed]
- Roth, J.S.; Juster, H. On the absence of ribonuclease inhibitor in rat liver nuclei. Biochim. Biophys. Acta Nucleic Acids Protein Synth. 1972, 287, 474–476. [Google Scholar] [CrossRef]
- Furia, A.; Moscato, M.; Calì, G.; Pizzo, E.; Confalone, E.; Amoroso, M.R.; Esposito, F.; Nitsch, L.; D’Alessio, G. The ribonuclease/angiogenin inhibitor is also present in mitochondria and nuclei. FEBS Lett. 2011, 585, 613–617. [Google Scholar] [CrossRef] [Green Version]
- Dickson, K.A.; Haigis, M.C.; Raines, R.T. Ribonuclease Inhibitor: Structure and Function. Prog. Nucleic Acid Res. Mol. Biol. 2005, 80, 349–374. [Google Scholar] [CrossRef] [Green Version]
- Smirnoff, P.; Roiz, L.; Angelkovitch, B.; Schwartz, B.; Shoseyov, O. A recombinant human RNASET2 glycoprotein with antitumorigenic and antiangiogenic characteristics. Cancer 2006, 107, 2760–2769. [Google Scholar] [CrossRef]
- Roiz, L.; Smirnoff, P.; Lewin, I.; Shoseyov, O.; Schwartz, B. Human recombinant RNASET2: A potential anti-cancer drug. Oncoscience 2016, 3, 71–84. [Google Scholar] [CrossRef] [Green Version]
- Acquati, F.; Mortara, L.; De Vito, A.; Baci, D.; Albini, A.; Cippitelli, M.; Taramelli, R.; Noonan, D.M. Innate Immune Response Regulation by the Human RNASET2 Tumor Suppressor Gene. Front. Immunol. 2019, 10, 2587. [Google Scholar] [CrossRef] [Green Version]
- Mastronicola, M.R.; Piccoli, R.; D’Alessio, G. Key Extracellular and Intracellular Steps in the Antitumor Action of Seminal Ribonuclease. JBIC J. Biol. Inorg. Chem. 1995, 230, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Tatsuta, T.; Hosono, M.; Sugawara, S.; Kariya, Y.; Ogawa, Y.; Hakomori, S.; Nitta, K. Sialic acid-binding lectin (leczyme) induces caspase-dependent apoptosis-mediated mitochondrial perturbation in Jurkat cells. Int. J. Oncol. 2013, 43, 1402–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iordanov, M.S.; Ryabinina, O.P.; Wong, J.; Dinh, T.H.; Newton, D.L.; Rybak, S.M.; Magun, B.E. Molecular determinants of apoptosis induced by the cytotoxic ribonuclease onconase: Evidence for cytotoxic mechanisms different from inhibition of protein synthesis. Cancer Res. 2000, 60, 1983–1994. [Google Scholar] [PubMed]
- Juan, G.; Ardelt, B.; Li, X.; Mikulski, S.M.; Shogen, K.; Ardelt, W.; Mittelman, A.; Darzynkiewicz, Z. G1 arrest of U937 cells by onconase is associated with suppression of cyclin D3 expression, induction of p16INK4A, p21WAF1/CIP1 and p27KIP and decreased pRb phosphorylation. Leukemia 1998, 12, 1241–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, S.; Ardelt, B.; Hsieh, T.-C.; Darzynkiewicz, Z.; Shogen, K.; Wu, J. Treatment of Jurkat acute T-lymphocytic leukemia cells by onconase (Ranpirnase) is accompanied by an altered nucleocytoplasmic distribution and reduced expression of transcription factor NF-κB. Int. J. Oncol. 2004, 25, 1745–1752. [Google Scholar] [CrossRef]
- Saxena, S.K.; Sirdeshmukh, R.; Ardelt, W.; Mikulski, S.M.; Shogen, K.; Youle, R.J. Entry into Cells and Selective Degradation of tRNAs by a Cytotoxic Member of the RNase A Family. J. Biol. Chem. 2002, 277, 15142–15146. [Google Scholar] [CrossRef] [Green Version]
- Altomare, D.A.; Rybak, S.M.; Pei, J.; Maizel, J.V.; Cheung, M.; Testa, J.R.; Shogen, K. Onconase responsive genes in human mesothelioma cells: Implications for an RNA damaging therapeutic agent. BMC Cancer 2010, 10, 34. [Google Scholar] [CrossRef] [Green Version]
- Ardelt, B.; Ardelt, W.; Darzynkiewicz, Z. Cytotoxic Ribonucleases and RNA Interference (RNAi). Cell Cycle 2003, 2, 22–24. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Ardelt, B.; Ardelt, W.; Shogen, K.; Darzynkiewicz, Z. The cytotoxic ribonuclease onconase targets RNA interference (siRNA). Cell Cycle 2008, 7, 3258–3261. [Google Scholar] [CrossRef]
- Qiao, M.; Zu, L.-D.; He, X.-H.; Shen, R.-L.; Wang, Q.-C.; Liu, M.-F. Onconase downregulates microRNA expression through targeting microRNA precursors. Cell Res. 2012, 22, 1199–1202. [Google Scholar] [CrossRef] [Green Version]
- Iordanov, M.S.; Wong, J.; Newton, D.L.; Rybak, S.M.; Bright, R.K.; Flavell, R.A.; Davis, R.J.; Magun, B.E. Differential Requirement for the Stress-Activated Protein Kinase/c-Jun NH2-Terminal Kinase in RNA Damage-Induced Apoptosis in Primary and in Immortalized Fibroblasts. Mol. Cell Biol. Res. Commun. 2000, 4, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Grabarek, J.; Ardelt, B.; Du, L.; Darzynkiewicz, Z. Activation of Caspases and Serine Proteases during Apoptosis Induced by Onconase (Ranpirnase). Exp. Cell Res. 2002, 278, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Yong, J.; Liu, H.; Shi, Y.; Meinkoth, J.; Dreyfuss, G.; Yang, X. tRNA Binds to Cytochrome c and Inhibits Caspase Activation. Mol. Cell 2010, 37, 668–678. [Google Scholar] [CrossRef] [Green Version]
- Michaelis, M.; Cinatl, J.; Anand, P.; Rothweiler, F.; Kotchetkov, R.; Von Deimling, A.; Doerr, H.W.; Shogen, K.; Cinatl, J., Jr. Onconase induces caspase-independent cell death in chemoresistant neuroblastoma cells. Cancer Lett. 2007, 250, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Fiorini, C.; Cordani, M.; Gotte, G.; Picone, D.; Donadelli, M. Onconase induces autophagy sensitizing pancreatic cancer cells to gemcitabine and activates Akt/mTOR pathway in a ROS-dependent manner. Biochim. Biophys. Acta Bioenerg. 2015, 1853, 549–560. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Nino, M.E.; Vianale, G.; Sabo-Attwood, T.; Mutti, L.; Porta, C.; Heintz, N.; Mossman, B.T. Human mesothelioma cells exhibit tumor cell–specific differences in phosphatidylinositol 3-kinase/AKT activity that predict the efficacy of Onconase. Mol. Cancer Ther. 2005, 4, 835–842. [Google Scholar] [CrossRef] [Green Version]
- Goparaju, C.M.; Blasberg, J.D.; Volinia, S.; Palatini, J.; Ivanov, S.V.; Donington, J.S.; Croce, C.; Carbone, M.; Yang, H.; Pass, H.I. Onconase mediated NFKβ downregulation in malignant pleural mesothelioma. Oncogene 2011, 30, 2767–2777. [Google Scholar] [CrossRef] [Green Version]
- Nasu, M.; Carbone, M.; Gaudino, G.; Ly, B.H.; Bertino, P.; Shimizu, D.; Morris, P.; Pass, H.I.; Yang, H. Ranpirnase Interferes with NF- B Pathway and MMP9 Activity, Inhibiting Malignant Mesothelioma Cell Invasiveness and Xenograft Growth. Genes Cancer 2011, 2, 576–584. [Google Scholar] [CrossRef]
- Raineri, A.; Fasoli, S.; Campagnari, R.; Gotte, G.; Menegazzi, M. Onconase Restores Cytotoxicity in Dabrafenib-Resistant A375 Human Melanoma Cells and Affects Cell Migration, Invasion and Colony Formation Capability. Int. J. Mol. Sci. 2019, 20, 5980. [Google Scholar] [CrossRef] [Green Version]
- Trang, P.; Medina, P.P.; Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Homer, R.; Brown, D.; Bader, A.G.; Weidhaas, J.B.; et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene 2009, 29, 1580–1587. [Google Scholar] [CrossRef] [Green Version]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribó, M.; Benito, A.; Vilanova, M. Antitumor ribonucleases. In Ribonucleases; Nicholson, A.W., Temple, U., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 26, pp. 55–88. ISBN 978-3-642-21077-8. [Google Scholar]
- Bosch, M.; Benito, A.; Ribo, M.; Puig, T.; Beaumelle, B.; Vilanova, M. A Nuclear Localization Sequence Endows Human Pancreatic Ribonuclease with Cytotoxic Activity. Biochemistry 2004, 43, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Xu, Z. Three decades of research on angiogenin: A review and perspective. Acta Biochim. Biophys. Sin. 2015, 48, 399–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.; Beintema, J.J.; Zhang, J. The ribonuclease A superfamily of mammals and birds: Identifying new members and tracing evolutionary histories. Genomics 2005, 85, 208–220. [Google Scholar] [CrossRef]
- Sorrentino, S. The eight human “canonical” ribonucleases: Molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 2010, 584, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-H.; Wang, Y.-N.; Hung, M.-C. Functional roles of the human ribonuclease A superfamily in RNA metabolism and membrane receptor biology. Mol. Asp. Med. 2019, 70, 106–116. [Google Scholar] [CrossRef]
- Shapiro, R.; Vallee, B.L. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin. Alanine derivatives inhibit angiogenin-induced angiogenesis. Biochemistry 1989, 28, 7401–7408. [Google Scholar] [CrossRef]
- Hu, G.F.; Strydom, D.J.; Fett, J.W.; Riordan, J.F.; Vallee, B.L. Actin is a binding protein for angiogenin. Proc. Natl. Acad. Sci. USA 1993, 90, 1217–1221. [Google Scholar] [CrossRef] [Green Version]
- Skorupa, A.; King, M.A.; Aparicio, I.M.; Dussmann, H.; Coughlan, K.; Breen, B.; Kieran, D.; Concannon, C.G.; Marin, P.; Prehn, J.H.M. Motoneurons Secrete Angiogenin to Induce RNA Cleavage in Astroglia. J. Neurosci. 2012, 32, 5024–5038. [Google Scholar] [CrossRef]
- Hu, G.-F.; Riordan, J.F.; Vallee, B.L. A putative angiogenin receptor in angiogenin-responsive human endothelial cells. Proc. Natl. Acad. Sci. USA 1997, 94, 2204–2209. [Google Scholar] [CrossRef] [Green Version]
- Yuxiang, J.; Goncalves, K.A.; Hailing, Y.; Kishikawa, H.; Sun, G.; Yang, H.; Vanli, N.; Wu, Y.; Jiang, Y.; Hu, M.G.; et al. Plexin-B2 Mediates Physiologic and Pathologic Functions of Angiogenin. Cell 2017, 171, 849–864.e25. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-N.; Lee, H.-H.; Chou, C.-K.; Yang, W.-H.; Wei, Y.; Chen, C.-T.; Yao, J.; Hsu, J.L.; Zhu, C.; Ying, H.; et al. Angiogenin/Ribonuclease 5 Is an EGFR Ligand and a Serum Biomarker for Erlotinib Sensitivity in Pancreatic Cancer. Cancer Cell 2018, 33, 752–769.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, J.; Hu, G.-F.; Xu, Z. Abstract 1854: The regulation of angiogenin expression and the genes regulated by angiogenin. Mol. Cell. Biol. 2016, 76, 1854. [Google Scholar] [CrossRef]
- Vert, A.; Castro, J.; Ribó, M.; Benito, A.; Vilanova, M. Activating transcription factor 3 is crucial for antitumor activity and to strengthen the antiviral properties of Onconase. Oncotarget 2016, 8, 11692–11707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brand, R.M.; Siegel, A.; Myerski, A.; Metter, E.J.; Engstrom, J.; Brand, R.E.; Squiquera, L.; Hodge, T.; Sulley, J.; Cranston, R.D.; et al. Ranpirnase Reduces HIV-1 Infection and Associated Inflammatory Changes in a Human Colorectal Explant Model. AIDS Res. Hum. Retrovir. 2018, 34, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.G.; Jackson, F.R.; Morgan, C.; Carson, W.C.; Martin, B.E.; Gallardo-Romero, N.; Ellison, J.A.; Greenberg, L.; Hodge, T.; Squiquera, L.; et al. Antiviral Ranpirnase TMR-001 Inhibits Rabies Virus Release and Cell-to-Cell Infection In Vitro. Viruses 2020, 12, 177. [Google Scholar] [CrossRef] [Green Version]
- Hodge, T.; Draper, K.; Brasel, T.; Freiberg, A.N.; Squiquera, L.; Sidransky, D.; Sulley, J.; Taxman, D.J. Antiviral effect of ranpirnase against Ebola virus. Antivir. Res. 2016, 132, 210–218. [Google Scholar] [CrossRef]
- Squiquera, L.; Taxman, D.J.; Brendle, S.A.; Torres, R.; Sulley, J.; Hodge, T.; Christensen, N.; Sidransky, D. Ranpirnase eradicates human papillomavirus in cultured cells and heals anogenital warts in a Phase I study. Antivir. Ther. 2017, 22, 247–255. [Google Scholar] [CrossRef]
- Mирoнoва, Н.Л.; Patutina, O.; Brenner, E.; Kurilshikov, A.; Vlassov, V.; Zenkova, M. The systemic tumor response to RNase A treatment affects the expression of genes involved in maintaining cell malignancy. Oncotarget 2017, 8, 78796–78810. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, M.; Benito, A.; Tubert, P.; Castro, J.; Ribo, M.; Beaumelle, B.; Vilanova, M. A Cytotoxic Ribonuclease Variant with a Discontinuous Nuclear Localization Signal Constituted by Basic Residues Scattered Over Three Areas of the Molecule. J. Mol. Biol. 2006, 360, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Vert, A.; Castro, J.; Ribo, M.; Benito, A.; Vilanova, M. A nuclear-directed human pancreatic ribonuclease (PE5) targets the metabolic phenotype of cancer cells. Oncotarget 2016, 7, 18309–18324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, Y.-F.; Chen, H.-H.; Lai, P.-F.; Cheng, C.-F.; Huang, Y.-T.; Lee, Y.-C.; Chen, T.-W.; Lin, H. MicroRNA-494 Reduces ATF3 Expression and Promotes AKI. J. Am. Soc. Nephrol. 2012, 23, 2012–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, J.; Ribó, M.; Benito, A.; Vilanova, M. Approaches to Endow Ribonucleases with Antitumor Activity: Lessons Learned from the Native Cytotoxic Ribonucleases. In Anti-Cancer Drugs—Nature, Synthesis and Cell; IntechOpen: London, UK, 2016; pp. 135–168. [Google Scholar]
- Maack, T.; Johnson, V.; Kau, S.T.; Figueiredo, J.; Sigulem, D. Renal filtration, transport, and metabolism of low-molecular-weight proteins: A review. Kidney Int. 1979, 16, 251–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costanzi, J.; Sidransky, D.; Navon, A.; Goldsweig, H. Ribonucleases as a Novel Pro-Apoptotic Anticancer Strategy: Review of the Preclinical and Clinical Data for Ranpirnase. Cancer Investig. 2005, 23, 643–650. [Google Scholar] [CrossRef]
- Vasandani, V.M.; Burris, J.A.; Sung, C. Reversible nephrotoxicity of onconase and effect of lysine pH on renal onconase uptake. Cancer Chemother. Pharmacol. 1999, 44, 164–169. [Google Scholar] [CrossRef]
- Vasandani, V.M.; Wu, Y.N.; Mikulski, S.M.; Youle, R.J.; Sung, C. Molecular determinants in the plasma clearance and tissue distribution of ribonucleases of the ribonuclease A superfamily. Cancer Res. 1996, 56, 4180–4186. [Google Scholar]
- Mikulski, S.; Grossman, A.; Carter, P.; Shogen, K.; Costanzi, J. Phase-i human clinical-trial of onconase(r) (p-30 protein) administered intravenously on a weekly schedule in cancer-patients with solid tumors. Int. J. Oncol. 1993, 3, 57–64. [Google Scholar] [CrossRef]
- Vogelzang, N.J.; Aklilu, M.; Stadler, W.M.; Dumas, M.C.; Mikulski, S.M. A phase II trial of weekly intravenous ranpirnase (Onconase), a novel ribonuclease in patients with metastatic kidney cancer. Investig. New Drugs 2001, 19, 255–260. [Google Scholar] [CrossRef]
- Chao, T.-Y.; Lavis, L.D.; Raines, R.T. Cellular Uptake of Ribonuclease A Relies on Anionic Glycans. Biochemistry 2010, 49, 10666–10673. [Google Scholar] [CrossRef] [Green Version]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Li, Q.; Neufeld, C.I.; Pouli, D.; Sun, S.; Yang, L.; Deng, P.; Wang, M.; Georgakoudi, I.; et al. Hyaluronic acid modification of RNase A and its intracellular delivery using lipid-like nanoparticles. J. Control. Release 2017, 263, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Jang, J.Y.; Joung, Y.K.; Kwon, M.H.; Park, K.D. Intracellular delivery and anti-cancer effect of self-assembled heparin-Pluronic nanogels with RNase A. J. Control. Release 2010, 147, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Kordalivand, N.; Li, D.; Beztsinna, N.; Toraño, J.S.; Mastrobattista, E.; Van Nostrum, C.F.; Hennink, W.E.; Vermonden, T. Polyethyleneimine coated nanogels for the intracellular delivery of RNase A for cancer therapy. Chem. Eng. J. 2018, 340, 32–41. [Google Scholar] [CrossRef]
- Khodzhaeva, V.; Makeeva, A.; Ulyanova, V.; Zelenikhin, P.; Evtugyn, V.; Hardt, M.; Rozhina, E.; Lvov, Y.; Fakhrullin, R.; Ilinskaya, O. Binase Immobilized on Halloysite Nanotubes Exerts Enhanced Cytotoxicity toward Human Colon Adenocarcinoma Cells. Front. Pharmacol. 2017, 8, 631. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Duan, F.; Liu, S.; Wu, T.; Shang, Y.; Tian, R.; Liu, J.; Wang, Z.-G.; Jiang, Q.; Ding, B. Efficient Intracellular Delivery of RNase A Using DNA Origami Carriers. ACS Appl. Mater. Interfaces 2019, 11, 11112–11118. [Google Scholar] [CrossRef]
- Wezler, X.; Dübel, S.; Schirrmann, T. Antibody fusion proteins with human ribonucleases 1 to 8. Hum. Antibodies 2018, 26, 177–192. [Google Scholar] [CrossRef]
- De Lorenzo, C.; D’Alessio, G. From ImmunoToxins to ImmunoRNases. Curr. Pharm. Biotechnol. 2008, 9, 210–214. [Google Scholar] [CrossRef]
- Schirrmann, T.; Krauss, J.; Arndt, M.A.; Rybak, S.M.; Dübel, S. Targeted therapeutic RNases (ImmunoRNases). Expert Opin. Biol. Ther. 2008, 9, 79–95. [Google Scholar] [CrossRef]
- Shramova, E.; Proshkina, G.; Shipunova, V.; Ryabova, A.; Kamyshinsky, R.; Konevega, A.; Schulga, A.; Konovalova, E.; Telegin, G.; Deyev, S.M. Dual Targeting of Cancer Cells with DARPin-Based Toxins for Overcoming Tumor Escape. Cancers 2020, 12, 3014. [Google Scholar] [CrossRef]
- Jordaan, S.; Akinrinmade, O.A.; Nachreiner, T.; Cremer, C.; Naran, K.; Chetty, S.; Barth, S. Updates in the Development of ImmunoRNases for the Selective Killing of Tumor Cells. Biomedicines 2018, 6, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nag, M.; Gajbhiye, V.; Kesharwani, P.; Jain, N.K. Transferrin functionalized chitosan-PEG nanoparticles for targeted delivery of paclitaxel to cancer cells. Colloids Surf. B Biointerfaces 2016, 148, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Ye, X.; Li, L.; Bai, H.; Xu, C. Improving the specific antitumor efficacy of ONC by fusion with N-terminal domain of transferrin. Biosci. Biotechnol. Biochem. 2018, 82, 1153–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima, F.R.; Kahn, S.A.; Soletti, R.C.; Biasoli, D.; Alves, T.; Da Fonseca, A.C.C.; Garcia, C.; Romão, L.; Brito, J.; Holanda-Afonso, R.; et al. Glioblastoma: Therapeutic challenges, what lies ahead. Biochim. Biophys. Acta Bioenerg. 2012, 1826, 338–349. [Google Scholar] [CrossRef]
- Kesavan, K.; Ratliff, J.; Johnson, E.W.; Dahlberg, W.; Asara, J.M.; Misra, P.; Frangioni, J.V.; Jacoby, D.B. Annexin A2 Is a Molecular Target for TM601, a Peptide with Tumor-targeting and Anti-angiogenic Effects. J. Biol. Chem. 2010, 285, 4366–4374. [Google Scholar] [CrossRef] [Green Version]
- Deshane, J.; Garner, C.C.; Sontheimer, H. Chlorotoxin Inhibits Glioma Cell Invasion via Matrix Metalloproteinase-2. J. Biol. Chem. 2003, 278, 4135–4144. [Google Scholar] [CrossRef] [Green Version]
- Mamelak, A.; Rosenfeld, S.; Bucholz, R.; Raubitschek, A.; Nabors, L.B.; Fiveash, J.B.; Shen, S.; Khazaeli, M.; Colcher, D.; Liu, A.; et al. Phase I Single-Dose Study of Intracavitary-Administered Iodine-131-TM-601 in Adults with Recurrent High-Grade Glioma. J. Clin. Oncol. 2006, 24, 3644–3650. [Google Scholar] [CrossRef]
- Mamelak, A.N.; Jacoby, D.B. Targeted delivery of antitumoral therapy to glioma and other malignancies with synthetic chlorotoxin (TM-601). Expert Opin. Drug Deliv. 2007, 4, 175–186. [Google Scholar] [CrossRef]
- Wang, X.; Guo, Z. Chlorotoxin-conjugated onconase as a potential anti-glioma drug. Oncol. Lett. 2015, 9, 1337–1342. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhang, P.; He, D.; Rödl, W.; Preiß, T.; Rädler, J.O.; Wagner, E.R.; Lächelt, U. pH-Reversible Cationic RNase A Conjugates for Enhanced Cellular Delivery and Tumor Cell Killing. Biomacromolecules 2015, 17, 173–182. [Google Scholar] [CrossRef]
- Rutkoski, T.J.; Kink, J.A.; Strong, L.E.; Raines, R.T. Site-specific PEGylation endows a mammalian ribonuclease with antitumor activity. Cancer Biol. Ther. 2011, 12, 208–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Shen, S.; Wen, D.; Li, M.; Li, T.; Chen, X.; Gu, Z.; Mo, R. Hierarchical Nanoassemblies-Assisted Combinational Delivery of Cytotoxic Protein and Antibiotic for Cancer Treatment. Nano Lett. 2018, 18, 2294–2303. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Tang, X.; Yang, J.; Zhang, J.; Han, H.; Li, Q. A genipin-crosslinked protein-polymer hybrid system for the intracellular delivery of ribonuclease A. Int. J. Nanomed. 2019, 14, 7389–7398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nel, A.E.; Mädler, L.; Velegol, D.; Xia, T.; Hoek, E.M.V.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M.O. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef]
- Behr, J.-P. The Proton Sponge: A Trick to Enter Cells the Viruses Did Not Exploit. CHIMIA Int. J. Chem. 1997, 51, 34–36. [Google Scholar]
- Ressler, V.T.; Mix, K.A.; Raines, R.T. Esterification Delivers a Functional Enzyme into a Human Cell. ACS Chem. Biol. 2019, 14, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Padrosa, D.R.; Castro, J.; Romero-Casañas, A.; Ribo, M.; Vilanova, M.; Benito, A. Construction of Highly Stable Cytotoxic Nuclear-Directed Ribonucleases. Molecules 2018, 23, 3273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vert, A.; Castro, J.; Ruiz-Martínez, S.; Tubert, P.; Escribano, D.; Ribo, M.; Vilanova, M.; Benito, A. Generation of New Cytotoxic Human Ribonuclease Variants Directed to the Nucleus. Mol. Pharm. 2012, 9, 2894–2902. [Google Scholar] [CrossRef]

| Nanocarrier System | Targeted Receptor | RNase | Action Mechanism | Ref |
|---|---|---|---|---|
| Hyaluronic acid lipid-based NP | CD-44 | RNase A | Tumor cell targeting Decrease excretion | [115] |
| Heparin-Pluronic nanogel | NS 1 | RNase A | Protect against protease degradation Decrease excretion | [116] |
| Methacrylate-derivatized anionic dextran reversed by coating it with polyethyleneimine | NS | RNase A | Enhance its intracellular delivery Decrease excretion | [117] |
| PLG-g-mPEG | NS | RNase A | Enhance cellular uptake under hypoxic conditions Decrease excretion | [1] |
| Halloysite nanotubes | NS | Binase | Improve cellular uptake and release Decrease excretion | [118] |
| DNA origami-based nanoplatform | NS | RNase A | Enhance uptake efficiency Decrease excretion | [119] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro, J.; Ribó, M.; Vilanova, M.; Benito, A. Strengths and Challenges of Secretory Ribonucleases as AntiTumor Agents. Pharmaceutics 2021, 13, 82. https://doi.org/10.3390/pharmaceutics13010082
Castro J, Ribó M, Vilanova M, Benito A. Strengths and Challenges of Secretory Ribonucleases as AntiTumor Agents. Pharmaceutics. 2021; 13(1):82. https://doi.org/10.3390/pharmaceutics13010082
Chicago/Turabian StyleCastro, Jessica, Marc Ribó, Maria Vilanova, and Antoni Benito. 2021. "Strengths and Challenges of Secretory Ribonucleases as AntiTumor Agents" Pharmaceutics 13, no. 1: 82. https://doi.org/10.3390/pharmaceutics13010082