Harnessing the Complete Repertoire of Conventional Dendritic Cell Functions for Cancer Immunotherapy
Abstract
:1. Introduction
2. The Enigmatic Role of Plasmacytoid DCs in the DC Continuum
| Subpopulation | Species | Marker | Level of Detection | Literature |
| cDC1 | Mouse | CD8α (splenic, resident cDC1) | Protein; RNA | [80,86] |
| CD1d | Protein | [86] | ||
| CD207; Langerin | Protein; RNA | [35,37,80,82] | ||
| CD24 | Protein | [30,35,82] | ||
| CD103+CD11b- migratory cDC1 | Protein | [35,42] | ||
| Shared | XCR1 | Protein; RNA | [30,33,42,82,86,87,88,89] | |
| CADM1; TSLC1; NECL-2; IGSF4; SynCAM1; | Protein; RNA | [30,31,33,42,82,86,87,88,89] | ||
| CLEC9A; DNGR-1; CD370 | Protein; RNA | [24,28,29,31,33,41,42,46,80,82,87,89,90,91] | ||
| BTLA | Protein; RNA | [30,40,42,86,90] | ||
| CD26 | Protein | [29,30,90] | ||
| Sirpα; CD172a negative | Protein | [28,30,92] | ||
| CD11c (human cDC1int) | Protein | [28,30,31,42,80,86,88] | ||
| MHC-II; HLA-DR | Protein | [28,29,30,42,86] | ||
| Human | CD141; BDCA-3 | Protein | [28,31,34,82,87,90,92] | |
| CD103+Sirpα- migratory cDC1 | Protein | [42] | ||
| cDC2 | Mouse | CD4 (lymphoid tissue) | Protein; RNA | [80,93] |
| DCIR2 (lymphoid tissue) | Protein; RNA | [55] | ||
| CD103+CD11b+ gut migratory cDC2 | Protein | [42] | ||
| Shared | Sirpα; CD172a | Protein | [28,29,30,31,35,42,80,82,86,88,92] | |
| XCR1 negative | Protein | [30] | ||
| CD11b | Protein; RNA | [28,30,42,80,88] | ||
| CD11c | Protein; RNA | [28,30,31,42,80,86,88] | ||
| MHC-II; HLA-DR | Protein | [28,29,30,42,86] | ||
| Human | CD1c; BDCA-1 | Protein; RNA | [28,31,34,80,90,91,92] | |
| CD1a (skin) | Protein | [28,94] | ||
| CLEC10A | Protein; RNA | [33,45] | ||
| CD163 | Protein | [90,92] | ||
| CD1d | Protein; RNA | [33,90] | ||
| FcεRIα | Protein; RNA | [33,80,90] | ||
| CD103+Sirpα+ gut migratory cDC2 | Protein | [42] | ||
| Subpopulation | Species | TLRs | Level of Detection | Literature |
| cDC1 | Mouse | TLR2 | Protein | [86] |
| TLR4; CD284 | RNA | [80,89] | ||
| TLR11 | RNA | [24,46,80,89] | ||
| TLR12 | RNA | [24,46,80,86,88,95] | ||
| TLR13 | Protein; RNA | [24,46,80,88] | ||
| CD180; RP105 | Protein | [86] | ||
| Shared | TLR3 | Protein; RNA | [42,80,87,88,89,92,96,97] | |
| TLR9 | Protein; RNA | [24,46,88,89,98] | ||
| Human | TLR6 | RNA | [89] | |
| TLR10; CD290 | Protein | [90] | ||
| cDC2 | Mouse | TLR9 | Protein; RNA | [24,46,80,88,98] |
| TLR13 | Protein; RNA | [24,46,88] | ||
| Shared | TLR1 | Protein; RNA | [80,86,89] | |
| TLR2; CD282 | Protein; RNA | [31,80,86,89,92] | ||
| TLR4; CD284 | RNA | [80,89] | ||
| TLR5 | RNA | [80,89,96,98] | ||
| TLR6 | Protein; RNA | [80,86,89] | ||
| TLR7 | Protein; RNA | [24,46,80,88,89,91] | ||
| TLR8 | Protein; RNA | [80,89,91,92] | ||
| CD180; RP105 | Protein | [86,90,92] | ||
| Human | - | - | - | |
| Subpopulation | Species | CLRs | Level of Detection | Literature |
| cDC1 | Mouse | CD207; Langerin | Protein | [35,86] |
| CLEC2D | Protein | [86] | ||
| CLEC4A; DCIR; CD367 | RNA | [42] | ||
| Shared | DEC205; CD205; Ly75 | Protein; RNA | [24,31,35,42,46,55,82,86,88,90] | |
| CLEC1A | RNA | [42] | ||
| CLEC9A; DNGR-1; CD370 | Protein; RNA | [24,28,29,31,33,41,42,46,82,87,89,91] | ||
| CLEC12A; MICL; KLRL1; CLL1 | Protein; RNA | [24,46,86,99] | ||
| Human | CD206; MMR (skin) | Protein | [28] | |
| Dectin-1; CLEC7A; CLECSF12; CD369 | Protein | [28] | ||
| cDC2 | Mouse | Dectin-2; CLEC6A; CLEC4N; CLECSF10 | Protein; RNA | [55,88] |
| CLEC4a2; DCIR1; CLECSF6 | RNA | [55,80,96] | ||
| CLEC4a3; DCIR3 | RNA | [55] | ||
| CLEC4a4; DCIR2 | Protein; RNA | [24,46,55,86,88] | ||
| DCAR | RNA | [55] | ||
| Shared | CLEC4A; DCIR; CD367 (subtype not defined) | Protein; RNA | [33,42,80,86,96] | |
| Dectin-1; CLEC7A; CLECSF12; CD369 | Protein; RNA | [28,55,86] | ||
| CLEC12A | Protein | [80,100] | ||
| CLEC13A; CD302 | RNA | [96] | ||
| CD209 (human gut); CD209a (mouse); DC-SIGN | Protein; RNA | [24,42,46,55,80,89,91] | ||
| Human | CD206; MMR (skin) | Protein | [28,42,90] | |
| CD207; Langerin | Protein; RNA | [42,91,94] | ||
| CLEC5A (thymic cDC2) | RNA | [31] | ||
| CLEC10A | Protein; RNA | [31,33,42,45,89] | ||
| CLEC11A; CLECSF3 (CD5 low cDC2) | RNA | [91] | ||
| CLEC17A (CD5 high cDC2) | RNA | [91] | ||
| Subpopulation | Species | FcγRs | Level of Detection | Literature |
| cDC1 | Mouse | FcγRIII; CD16 | Protein; RNA | [56,80,96] |
| FcγRIV; CD16.2 | Protein | [56] | ||
| Shared | FcγRI; CD64 | Protein; RNA | [30,42,56] | |
| FcγRIIB; CD32b | Protein; RNA | [56,80,86,101] | ||
| Human | FcγRIIA; CD32 | RNA | [102] | |
| cDC2 | Mouse | FcγRIV; CD16.2 | Protein | [56] |
| FcγRI; CD64 negative | Protein; RNA | [30,56,80] | ||
| Shared | FcγRIIB; CD32b | Protein; RNA | [33,42,56,80,86,89,102] | |
| FcγRIII; CD16; human FcγRIIIA | Protein; RNA | [80,101,102] | ||
| Human | FcγRI; CD64 | Protein; RNA | [89,101] | |
| FcγRIIA; CD32 | Protein; RNA | [28,31,96,101,102] | ||
| Subpopulation | Species | Intracellular Sensors | Level of Detection | Literature |
| cDC1 | Mouse | NLRP3; NALP3; Cryopyrin | Protein | [88] |
| Shared | ||||
| Human | NLRC5 | RNA | [42] | |
| Caspase-1 low | RNA | [92] | ||
| AIM2 | RNA | [92] | ||
| PYCARD | RNA | [92] | ||
| cDC2 | Mouse | NOD-1; NLRC1 | Protein; RNA | [24,46,88] |
| CARD9 | Protein | [88] | ||
| STING | Protein | [88] | ||
| MDA5 | Protein | [88] | ||
| RIG-I | Protein | [88] | ||
| NLRC4 | RNA | [24,46] | ||
| Shared | NLRP3; NALP3; Cryopyrin | Protein; RNA | [33,42,88,92] | |
| Caspase-1 high | Protein; RNA | [42,88,92] | ||
| Human | Caspase-8 | RNA | [92] | |
| NAIP | RNA | [92] | ||
| NLRC4 | RNA | [92] | ||
| NLRP1 | RNA | [92] | ||
| PYCARD | RNA | [92] | ||
| Subpopulation | Species | Co-Regulatory Molecules | Level of Detection | Literature |
| cDC1 | Mouse | - | - | - |
| Shared | CD80 | Protein | [30,42,55,82,87] | |
| CD86 | Protein; RNA | [31,33,42,55,80,82,87,89,96] | ||
| CD40 | Protein; RNA | [30,42,55,89] | ||
| PD-L1; CD274 | Protein | [28,30,82,87,89] | ||
| Human | IDO-1 | Protein; RNA | [31,33,42,80,89,92] | |
| IDO-2 | RNA | [42] | ||
| cDC2 | Mouse | - | - | - |
| Shared | CD80 | Protein | [30,42,55,82,87] | |
| CD86 | Protein; RNA | [30,31,33,42,55,82,87,96] | ||
| CD40 | Protein | [30,42,55,89] | ||
| PD-L1; CD274 | Protein | [30,80,82,87,89] | ||
| Human | ICOS-L | RNA | [28] | |
| Subpopulation | Species | Other Interesting Molecules | Level of Detection | Literature |
| cDC1 | Mouse | CD36 (Scavenger receptor) | Protein | [86,88] |
| Shared | CD135; FLT3 | Protein; RNA | [28,80,91] | |
| Human | TIM-3 | Protein | [90] | |
| cDC2 | Mouse | Clec12A (marker for cDC2B) | Protein | [80] |
| Clec10A (marker for cDC2B) | Protein | [80] | ||
| ESAM (marker for cDC2A or Notch2-dependent cDC2) | Protein | [66,68,80] | ||
| Shared | CD135; FLT3 | Protein; RNA | [28,80,91] | |
| Human | CD36 (marker for DC3; DC2 negative) | RNA | [31,33] | |
| CD163 (marker for DC3; DC2 negative) | Protein; RNA | [28,33] | ||
| CD5 (marker for DC2; DC3 negative) | Protein; RNA | [90,91] | ||
| CD200R (downregulating DC activity) | Protein | [90] |
3. Environmental Cues Controlling the Stimulatory and Tolerogenic Capacity of DCs
3.1. Dendritic Cells Function as Sensors for Invading Pathogens
3.2. Pathogen Recognition Receptors on DCs Regulate Antigen Sensing, Uptake and Activation
| TLR Ligands | ||||
| Receptor | Organism | Ligand Class | Molecules | References |
| TLR2 (TLR1/TLR2; TLR2/TLR6) | M/H | Pathogenic | Lipoptroteins; Triacetylated lipopeptides; human β-defensin 3 (BD-3); Glycosylphosphatidylinositol (GPI); Zymosan; Peptidoglycan; LPS; Lipoproteine/-peptide; Glycolipide; MALP-2; Diacetylierte Lipopeptide; LTA; Zymosan | [244,245,246,247] [248,249,250,251,252,253,254,255,256,257,258] |
| Endogenous | Endoplasmin; Hsp60; Hsp70; Human cardiac myosin; Urate crystals; Hyaluronan | [259,260,261,262,263,264] | ||
| Synthetic | FSL-1, synthetic lipopeptides & lipoprotein analogs, triacetylated lipopeptides e.g., Pam2CSK4, Pam3CSK4, or synthetic beta-defensin 3, MALP2 | [244,245,258], [265,266,267,268] | ||
| TLR3 | M/H | Pathogenic | dsRNA | [269,270,271] |
| Endogenous | mRNA | [270] | ||
| Synthetic | poly(I:C); poly(A:U) | [258,272,273,274] | ||
| TLR4 (MD-2; CD14) | M/H | Pathogenic | Lipopolysaccharid (LPS; Gram-); Lipoteichoic acid (LTA; Gram+) | [256,275,276] |
| Endogenous | β-defensin 2; Fibronectin; Fibrinogen; Hyaluronan; Surfactant protein A; Urate crystals; OxPAPC; Hsp72; Hsp70; Hsp60; HMGB1; Endoplasmin | [262,264,277,278,279,280,281,282], [259,260,283,284] | ||
| Synthetic | Glycan-based agonists; Monophosphoryl Lipid A (MPL); pyrimido[5,4-b]-indoles; LPS | [268,285,286,287] | ||
| TLR5 | M/H | Pathogenic | Flagellin | [248,288] |
| Endogenous | Unknown | |||
| Synthetic | Flagellin | [289,290] | ||
| TLR7 | M/H | Pathogenic | ssRNA | [291,292,293] |
| Endogenous | RNA; siRNA | [294,295] | ||
| Synthetic | Imiquimod (R837); Resiquimod (R848); Gardiquimod; Loxoribine | [268,274,296,297,298,299,300,301] | ||
| TLR8 | M/H | Pathogenic | TLR7 antagonist (mouse); ssRNA (human) | [292,299,302,303] |
| Endogenous | Human cardiac myosin; siRNA | [295,304] | ||
| Synthetic | Imiquimod (R837); Resiquimod (R848); Gardiquimod; TL8-506 | [268,274,296,297,298,299,300,301] | ||
| TLR9 | M/H | Pathogenic | Unmethylated CpG DNA; dsDNA | [258,269,305,306] |
| Endogenous | DNA; HMGB1 | [307,308,309] | ||
| Synthetic | Diverse synthetic CpG-oligonucleotides (CpG-ODNs) | [268,274,310,311,312] | ||
| TLR10 | H | Pathogenic | Non functional in mice (viral insertion); TLR2 antagonist; HIV-1 gp41 | [120,313,314] |
| Endogenous | Unknown | |||
| Synthetic | Unknown | |||
| TLR11 | M | Pathogenic | Profilin | [95,120,315] |
| Endogenous | Unknown | |||
| Synthetic | Unknown | |||
| TLR12 | M | Pathogenic | Profilin | [120,315,316] |
| Endogenous | Unknown | |||
| Synthetic | Unknown | |||
| TLR13 | M | Pathogenic | Bacterial 23S rRNA | [120,123] |
| Endogenous | Unknown | |||
| Synthetic | Unknown | |||
| CD180 | M/H | Pathogenic | Unknown | |
| Endogenous | Negative regulator of TLR7 & TLR9 | [317] | ||
| Synthetic | Unknown | |||
| STING ligands | Organism | Ligand Class | Molecules | Literature |
| Receptor | ||||
| STING | M/H | Pathogenic | Bacterial cyclic di-nucleotides and cGAMP; viral DNA | [111,206,210,211] |
| Endogenous | Self-DNA e.g., dead cells, DNA leaking into the cytosol following stress or tumor DNA; tumor cGAMP | [220,225,227,228] | ||
| Synthetic | Cyclic dinucleotides e.g., 2′-3′-cGAMP; c-di-AMP; di-amidobenzimidazole | [301,318] | ||
| RLR ligands | Organism | Ligand Class | Molecules | Literature |
| Receptor | ||||
| RIG-I | M/H | Pathogenic | Paramyxoviridae, Rhabdoviridae & Orthomyxoviridae; short dsRNA; dsDNA in cooperation with RNA Polymerase III; 5′-phosphorylated ssRNAs | [110,155,158,160,319] |
| Endogenous | Endogeneous RNAs e.g., LINE1 | [320,321] | ||
| Synthetic | poly(dA:dT); tri-phosphorylated 5′ stem-loop RNAs | [158,322,323] | ||
| MDA-5 | M/H | Pathogenic | Picornaviridae; long dsRNA; ssRNA; dsDNA; NAB2; rb-dsRNA | [110,155,160,322,324] |
| Endogenous | Endogeneous RNAs e.g., mitochondrial RNA and retroelement transcripts | [320,325,326,327] | ||
| Synthetic | poly(I:C) | [160,322] | ||
4. DCs Orchestrate T Cell-Driven Immunity
4.1. The Potential of DC:T Cell-Based Vaccines and the Importance of CD4+ T Cell Help for Cancer Immunotherapy
4.2. The Priming of Naïve T Cells by DCs Is Regulated by Three Signals
4.2.1. Signal 1: pMHC Recognition by the TCR
4.2.2. Signal 2: Co-Regulatory Surface Molecules Define the Functional Phenotype of Primed T Cells
4.2.3. Signal 3: DC Cytokine Secretion Determines T Cell Polarization
5. DCs in the Tumor Microenvironment: Tipping the Scales during Anti-Cancer Immune Responses?
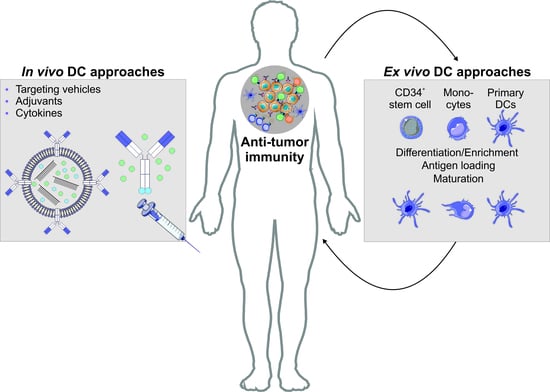
6. Harnessing the Potential of the Immune Systems’ Generals during Immunotherapeutic Approaches
6.1. Autologous DC Transfer Approaches for the Treatment of Malignancies
6.2. Antigen Targeting Enables the Induction of Antigen Specific T Cell Effector Modules Following Antigen Delivery to DCs In Vivo
6.2.1. The Principles of Antigen Delivery to DCs Utilizing Antigen-Coupled Targeting Antibodies In Vivo
6.2.2. Engineering of Antigen-Coupled Targeting Antibodies and Antibody-Coated Transport Vehicles
6.2.3. Factors Contributing in Rationale for DC Vaccine Design
7. Combination Therapies—Potential and Hurdles
8. Summary of DC-Based Immunotherapeutic Approaches Unleashing Anti-Tumor Immunity
9. Single Cell Omics Reshape Our Understanding of DC Biology Providing New Strategies for DC-Based Immunotherapeutic Approaches
Author Contributions
Funding
Conflicts of Interest
References
- Amon, L.; Lehmann, C.H.K.; Baranska, A.; Schoen, J.; Heger, L.; Dudziak, D. Transcriptional control of dendritic cell development and functions. Int. Rev. Cell Mol. Boil. 2019, 349, 55–151. [Google Scholar] [CrossRef]
- Heidkamp, G.F.; Lehmann, C.H.K.; Heger, L.; Baransk, A.; Hoffmann, A.; Lühr, J.; Dudziak, D. Functional Specialization of Dendritic Cell Subsets. In Encyclopedia of Cell Biology; Bradshaw, R.A., Stahl, P.D., Eds.; Academic Press: Waltham, MA, USA, 2016; pp. 588–604. [Google Scholar] [CrossRef]
- Steinman, R.M.; Banchereau, J. Taking dendritic cells into medicine. Nature 2007, 449, 419–426. [Google Scholar] [CrossRef]
- Kaplan, D.H. Ontogeny and function of murine epidermal Langerhans cells. Nat. Immunol. 2017, 18, 1068–1075. [Google Scholar] [CrossRef]
- Flacher, V.; Tripp, C.H.; Mairhofer, D.G.; Steinman, R.M.; Stoitzner, P.; Idoyaga, J.; Romani, N. Murine Langerin+ dermal dendritic cells prime CD8+ T cells while Langerhans cells induce cross-tolerance. EMBO Mol. Med. 2014, 6, 1638. [Google Scholar] [CrossRef]
- Igyarto, B.Z.; Haley, K.; Ortner, D.; Bobr, A.; Gerami-Nejad, M.; Edelson, B.T.; Zurawski, S.M.; Malissen, B.; Zurawski, G.; Berman, J.; et al. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity 2011, 35, 260–272. [Google Scholar] [CrossRef] [Green Version]
- Levin, C.; Bonduelle, O.; Nuttens, C.; Primard, C.; Verrier, B.; Boissonnas, A.; Combadiere, B. Critical Role for Skin-Derived Migratory DCs and Langerhans Cells in TFH and GC Responses after Intradermal Immunization. J. Investing. Dermatol. 2017, 137, 1905–1913. [Google Scholar] [CrossRef] [Green Version]
- Mathers, A.R.; Janelsins, B.M.; Rubin, J.P.; Tkacheva, O.A.; Shufesky, W.J.; Watkins, S.C.; Morelli, A.E.; Larregina, A.T. Differential capability of human cutaneous dendritic cell subsets to initiate Th17 responses. J. Immunol. (Baltim. Md. 1950) 2009, 182, 921–933. [Google Scholar] [CrossRef] [Green Version]
- Stoitzner, P.; Tripp, C.H.; Eberhart, A.; Price, K.M.; Jung, J.Y.; Bursch, L.; Ronchese, F.; Romani, N. Langerhans cells cross-present antigen derived from skin. Proc. Natl. Acad. Sci. USA 2006, 103, 7783–7788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malissen, B.; Tamoutounour, S.; Henri, S. The origins and functions of dendritic cells and macrophages in the skin. Nat. Rev. Immunol. 2014, 14, 417–428. [Google Scholar] [PubMed]
- Segura, E.; Amigorena, S. Inflammatory dendritic cells in mice and humans. Trends Immunol. 2013, 34, 440–445. [Google Scholar] [CrossRef] [Green Version]
- Hammad, H.; Plantinga, M.; Deswarte, K.; Pouliot, P.; Willart, M.A.M.; Kool, M.; Muskens, F.; Lambrecht, B.N. Inflammatory dendritic cells—not basophils—are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J. Exp. Med. 2010, 207, 2097–2111. [Google Scholar] [CrossRef]
- Hohl, T.M.; Rivera, A.; Lipuma, L.; Gallegos, A.; Shi, C.; Mack, M.; Pamer, E.G. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe 2009, 6, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Leon, B.; Lopez-Bravo, M.; Ardavin, C. Monocyte-derived dendritic cells formed at the infection site control the induction of protective T helper 1 responses against Leishmania. Immunity 2007, 26, 519–531. [Google Scholar] [CrossRef] [Green Version]
- Segura, E.; Albiston, A.L.; Wicks, I.P.; Chai, S.Y.; Villadangos, J.A. Different cross-presentation pathways in steady-state and inflammatory dendritic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 20377–20381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakim, L.M.; Waithman, J.; van Rooijen, N.; Heath, W.R.; Carbone, F.R. Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science (N. Y.) 2008, 319, 198–202. [Google Scholar] [CrossRef] [Green Version]
- Bender, A.; Sapp, M.; Schuler, G.; Steinman, R.M.; Bhardwaj, N. Improved methods for the generation of dendritic cells from nonproliferating progenitors in human blood. J. Immunol. Methods 1996, 196, 121–135. [Google Scholar] [CrossRef]
- Lutz, M.B.; Kukutsch, N.; Ogilvie, A.L.; Rossner, S.; Koch, F.; Romani, N.; Schuler, G. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods 1999, 223, 77–92. [Google Scholar] [CrossRef]
- Helft, J.; Bottcher, J.; Chakravarty, P.; Zelenay, S.; Huotari, J.; Schraml, B.U.; Goubau, D.; Reis e Sousa, C. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCII(+) Macrophages and Dendritic Cells. Immunity 2015, 42, 1197–1211. [Google Scholar] [CrossRef] [Green Version]
- Guilliams, M.; Malissen, B. A Death Notice for In-Vitro-Generated GM-CSF Dendritic Cells? Immunity 2015, 42, 988–990. [Google Scholar] [CrossRef] [Green Version]
- Amon, L.; Lehmann, C.H.K.; Heger, L.; Heidkamp, G.F.; Dudziak, D. The ontogenetic path of human dendritic cells. Mol. Immunol. 2020, 120, 122–129. [Google Scholar] [CrossRef]
- Guilliams, M.; Ginhoux, F.; Jakubzick, C.; Naik, S.H.; Onai, N.; Schraml, B.U.; Segura, E.; Tussiwand, R.; Yona, S. Dendritic cells, monocytes and macrophages: A unified nomenclature based on ontogeny. Nat. Rev. Immunol. 2014, 14, 571–578. [Google Scholar] [CrossRef]
- Merad, M.; Ginhoux, F.; Collin, M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat. Rev. Immunol. 2008, 8, 935–947. [Google Scholar] [CrossRef] [PubMed]
- Merad, M.; Sathe, P.; Helft, J.; Miller, J.; Mortha, A. The dendritic cell lineage: Ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013, 31, 563–604. [Google Scholar] [CrossRef] [Green Version]
- Murphy, T.L.; Grajales-Reyes, G.E.; Wu, X.; Tussiwand, R.; Briseno, C.G.; Iwata, A.; Kretzer, N.M.; Durai, V.; Murphy, K.M. Transcriptional Control of Dendritic Cell Development. Annu. Rev. Immunol. 2016, 34, 93–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenbarth, S.C. Dendritic cell subsets in T cell programming: Location dictates function. Nat. Rev. Immunol. 2019, 19, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Worbs, T.; Hammerschmidt, S.I.; Forster, R. Dendritic cell migration in health and disease. Nat. Rev. Immunol. 2017, 17, 30–48. [Google Scholar] [CrossRef]
- Alcantara-Hernandez, M.; Leylek, R.; Wagar, L.E.; Engleman, E.G.; Keler, T.; Marinkovich, M.P.; Davis, M.M.; Nolan, G.P.; Idoyaga, J. High-Dimensional Phenotypic Mapping of Human Dendritic Cells Reveals Interindividual Variation and Tissue Specialization. Immunity 2017, 47, 1037–1050.e1036. [Google Scholar] [CrossRef] [Green Version]
- Granot, T.; Senda, T.; Carpenter, D.J.; Matsuoka, N.; Weiner, J.; Gordon, C.L.; Miron, M.; Kumar, B.V.; Griesemer, A.; Ho, S.H.; et al. Dendritic Cells Display Subset and Tissue-Specific Maturation Dynamics over Human Life. Immunity 2017, 46, 504–515. [Google Scholar] [CrossRef] [Green Version]
- Guilliams, M.; Dutertre, C.A.; Scott, C.L.; McGovern, N.; Sichien, D.; Chakarov, S.; Van Gassen, S.; Chen, J.; Poidinger, M.; De Prijck, S.; et al. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity 2016, 45, 669–684. [Google Scholar] [CrossRef] [Green Version]
- Heidkamp, G.F.; Sander, J.; Lehmann, C.H.K.; Heger, L.; Eissing, N.; Baranska, A.; Luhr, J.J.; Hoffmann, A.; Reimer, K.C.; Lux, A.; et al. Human lymphoid organ dendritic cell identity is predominantly dictated by ontogeny, not tissue microenvironment. Sci. Immunol. 2016, 1, eaai7677. [Google Scholar] [CrossRef]
- See, P.; Dutertre, C.A.; Chen, J.; Gunther, P.; McGovern, N.; Irac, S.E.; Gunawan, M.; Beyer, M.; Handler, K.; Duan, K.; et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science (N. Y.) 2017, 356. [Google Scholar] [CrossRef] [Green Version]
- Villani, A.C.; Satija, R.; Reynolds, G.; Sarkizova, S.; Shekhar, K.; Fletcher, J.; Griesbeck, M.; Butler, A.; Zheng, S.; Lazo, S.; et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science (N. Y.) 2017, 356. [Google Scholar] [CrossRef] [Green Version]
- Dzionek, A.; Fuchs, A.; Schmidt, P.; Cremer, S.; Zysk, M.; Miltenyi, S.; Buck, D.W.; Schmitz, J. BDCA-2, BDCA-3, and BDCA-4: Three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. (Baltim. Md. 1950) 2000, 165, 6037–6046. [Google Scholar] [CrossRef] [Green Version]
- Edelson, B.T.; Kc, W.; Juang, R.; Kohyama, M.; Benoit, L.A.; Klekotka, P.A.; Moon, C.; Albring, J.C.; Ise, W.; Michael, D.G.; et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 2010, 207, 823–836. [Google Scholar] [CrossRef] [Green Version]
- Everts, B.; Tussiwand, R.; Dreesen, L.; Fairfax, K.C.; Huang, S.C.; Smith, A.M.; O’Neill, C.M.; Lam, W.Y.; Edelson, B.T.; Urban, J.F., Jr.; et al. Migratory CD103+ dendritic cells suppress helminth-driven type 2 immunity through constitutive expression of IL-12. J. Exp. Med. 2016, 213, 35–51. [Google Scholar] [CrossRef]
- Ginhoux, F.; Collin, M.P.; Bogunovic, M.; Abel, M.; Leboeuf, M.; Helft, J.; Ochando, J.; Kissenpfennig, A.; Malissen, B.; Grisotto, M.; et al. Blood-derived dermal langerin+ dendritic cells survey the skin in the steady state. J. Exp. Med. 2007, 204, 3133–3146. [Google Scholar] [CrossRef]
- Henri, S.; Poulin, L.F.; Tamoutounour, S.; Ardouin, L.; Guilliams, M.; de Bovis, B.; Devilard, E.; Viret, C.; Azukizawa, H.; Kissenpfennig, A.; et al. CD207+ CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. J. Exp. Med. 2010, 207, 189–206. [Google Scholar] [CrossRef] [Green Version]
- Hildner, K.; Edelson, B.T.; Purtha, W.E.; Diamond, M.; Matsushita, H.; Kohyama, M.; Calderon, B.; Schraml, B.U.; Unanue, E.R.; Diamond, M.S.; et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science (N. Y.) 2008, 322, 1097–1100. [Google Scholar] [CrossRef] [Green Version]
- Jones, A.; Bourque, J.; Kuehm, L.; Opejin, A.; Teague, R.M.; Gross, C.; Hawiger, D. Immunomodulatory Functions of BTLA and HVEM Govern Induction of Extrathymic Regulatory T Cells and Tolerance by Dendritic Cells. Immunity 2016, 45, 1066–1077. [Google Scholar] [CrossRef] [Green Version]
- Sancho, D.; Joffre, O.P.; Keller, A.M.; Rogers, N.C.; Martinez, D.; Hernanz-Falcon, P.; Rosewell, I.; Reis e Sousa, C. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature 2009, 458, 899–903. [Google Scholar] [CrossRef]
- Watchmaker, P.B.; Lahl, K.; Lee, M.; Baumjohann, D.; Morton, J.; Kim, S.J.; Zeng, R.; Dent, A.; Ansel, K.M.; Diamond, B.; et al. Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat. Immunol. 2014, 15, 98–108. [Google Scholar] [CrossRef]
- Becker, M.; Guttler, S.; Bachem, A.; Hartung, E.; Mora, A.; Jakel, A.; Hutloff, A.; Henn, V.; Mages, H.W.; Gurka, S.; et al. Ontogenic, Phenotypic, and Functional Characterization of XCR1(+) Dendritic Cells Leads to a Consistent Classification of Intestinal Dendritic Cells Based on the Expression of XCR1 and SIRPalpha. Front. Immunol. 2014, 5, 326. [Google Scholar] [CrossRef] [Green Version]
- Flores-Langarica, A.; Cook, C.; Muller Luda, K.; Persson, E.K.; Marshall, J.L.; Beristain-Covarrubias, N.; Yam-Puc, J.C.; Dahlgren, M.; Persson, J.J.; Uematsu, S.; et al. Intestinal CD103(+)CD11b(+) cDC2 Conventional Dendritic Cells Are Required for Primary CD4(+) T and B Cell Responses to Soluble Flagellin. Front. Immunol. 2018, 9, 2409. [Google Scholar] [CrossRef] [PubMed]
- Heger, L.; Balk, S.; Luhr, J.J.; Heidkamp, G.F.; Lehmann, C.H.K.; Hatscher, L.; Purbojo, A.; Hartmann, A.; Garcia-Martin, F.; Nishimura, S.I.; et al. CLEC10A Is a Specific Marker for Human CD1c(+) Dendritic Cells and Enhances Their Toll-Like Receptor 7/8-Induced Cytokine Secretion. Front. Immunol. 2018, 9, 744. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.C.; Brown, B.D.; Shay, T.; Gautier, E.L.; Jojic, V.; Cohain, A.; Pandey, G.; Leboeuf, M.; Elpek, K.G.; Helft, J.; et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 2012, 13, 888–899. [Google Scholar] [CrossRef]
- Bosteels, C.; Neyt, K.; Vanheerswynghels, M.; van Helden, M.J.; Sichien, D.; Debeuf, N.; De Prijck, S.; Bosteels, V.; Vandamme, N.; Martens, L.; et al. Inflammatory Type 2 cDCs Acquire Features of cDC1s and Macrophages to Orchestrate Immunity to Respiratory Virus Infection. Immunity 2020, 52, 1039–1056.e1039. [Google Scholar] [CrossRef] [PubMed]
- Dzionek, A.; Sohma, Y.; Nagafune, J.; Cella, M.; Colonna, M.; Facchetti, F.; Gunther, G.; Johnston, I.; Lanzavecchia, A.; Nagasaka, T.; et al. BDCA-2, a novel plasmacytoid dendritic cell-specific type II C-type lectin, mediates antigen capture and is a potent inhibitor of interferon alpha/beta induction. J. Exp. Med. 2001, 194, 1823–1834. [Google Scholar] [CrossRef]
- Rissoan, M.C.; Duhen, T.; Bridon, J.M.; Bendriss-Vermare, N.; Peronne, C.; de Saint Vis, B.; Briere, F.; Bates, E.E. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood 2002, 100, 3295–3303. [Google Scholar] [CrossRef] [Green Version]
- Schlitzer, A.; Sivakamasundari, V.; Chen, J.; Sumatoh, H.R.; Schreuder, J.; Lum, J.; Malleret, B.; Zhang, S.; Larbi, A.; Zolezzi, F.; et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 2015, 16, 718–728. [Google Scholar] [CrossRef]
- Reizis, B. Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity 2019, 50, 37–50. [Google Scholar] [CrossRef] [Green Version]
- Schraml, B.U.; Reis e Sousa, C. Defining dendritic cells. Curr. Opin. Immunol. 2015, 32, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Den Haan, J.M.; Bevan, M.J. Constitutive versus activation-dependent cross-presentation of immune complexes by CD8(+) and CD8(-) dendritic cells in vivo. J. Exp. Med. 2002, 196, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Den Haan, J.M.; Lehar, S.M.; Bevan, M.J. CD8(+) but not CD8(-) dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 2000, 192, 1685–1696. [Google Scholar] [CrossRef] [PubMed]
- Dudziak, D.; Kamphorst, A.O.; Heidkamp, G.F.; Buchholz, V.R.; Trumpfheller, C.; Yamazaki, S.; Cheong, C.; Liu, K.; Lee, H.W.; Park, C.G.; et al. Differential antigen processing by dendritic cell subsets in vivo. Science (N. Y.) 2007, 315, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, C.H.K.; Baranska, A.; Heidkamp, G.F.; Heger, L.; Neubert, K.; Luhr, J.J.; Hoffmann, A.; Reimer, K.C.; Bruckner, C.; Beck, S.; et al. DC subset-specific induction of T cell responses upon antigen uptake via Fcgamma receptors in vivo. J. Exp. Med. 2017, 214, 1509–1528. [Google Scholar] [CrossRef]
- Idoyaga, J.; Lubkin, A.; Fiorese, C.; Lahoud, M.H.; Caminschi, I.; Huang, Y.; Rodriguez, A.; Clausen, B.E.; Park, C.G.; Trumpfheller, C.; et al. Comparable T helper 1 (Th1) and CD8 T-cell immunity by targeting HIV gag p24 to CD8 dendritic cells within antibodies to Langerin, DEC205, and Clec9A. Proc. Natl. Acad. Sci. USA 2011, 108, 2384–2389. [Google Scholar] [CrossRef] [Green Version]
- Kano, S.; Sato, K.; Morishita, Y.; Vollstedt, S.; Kim, S.; Bishop, K.; Honda, K.; Kubo, M.; Taniguchi, T. The contribution of transcription factor IRF1 to the interferon-gamma-interleukin 12 signaling axis and TH1 versus TH-17 differentiation of CD4+ T cells. Nat. Immunol. 2008, 9, 34–41. [Google Scholar] [CrossRef]
- Maldonado-Lopez, R.; Maliszewski, C.; Urbain, J.; Moser, M. Cytokines regulate the capacity of CD8alpha(+) and CD8alpha(-) dendritic cells to prime Th1/Th2 cells in vivo. J. Immunol. (Baltim. Md. 1950) 2001, 167, 4345–4350. [Google Scholar] [CrossRef] [Green Version]
- Wilson, D.C.; Matthews, S.; Yap, G.S. IL-12 signaling drives CD8+ T cell IFN-gamma production and differentiation of KLRG1+ effector subpopulations during Toxoplasma gondii Infection. J. Immunol. (Baltim. Md. 1950) 2008, 180, 5935–5945. [Google Scholar] [CrossRef] [Green Version]
- Bonifaz, L.; Bonnyay, D.; Mahnke, K.; Rivera, M.; Nussenzweig, M.C.; Steinman, R.M. Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J. Exp. Med. 2002, 196, 1627–1638. [Google Scholar] [CrossRef]
- Hawiger, D.; Inaba, K.; Dorsett, Y.; Guo, M.; Mahnke, K.; Rivera, M.; Ravetch, J.V.; Steinman, R.M.; Nussenzweig, M.C. Dendritic cells induce peripheral T cell unresponsiveness under steady state conditions in vivo. J. Exp. Med. 2001, 194, 769–779. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, S.; Dudziak, D.; Heidkamp, G.F.; Fiorese, C.; Bonito, A.J.; Inaba, K.; Nussenzweig, M.C.; Steinman, R.M. CD8+ CD205+ splenic dendritic cells are specialized to induce Foxp3+ regulatory T cells. J. Immunol. (Baltim. Md. 1950) 2008, 181, 6923–6933. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Nish, S.A.; Jiang, R.; Hou, L.; Licona-Limon, P.; Weinstein, J.S.; Zhao, H.; Medzhitov, R. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity 2013, 39, 722–732. [Google Scholar] [CrossRef] [Green Version]
- Kinnebrew, M.A.; Buffie, C.G.; Diehl, G.E.; Zenewicz, L.A.; Leiner, I.; Hohl, T.M.; Flavell, R.A.; Littman, D.R.; Pamer, E.G. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity 2012, 36, 276–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, K.L.; Caton, M.L.; Bogunovic, M.; Greter, M.; Grajkowska, L.T.; Ng, D.; Klinakis, A.; Charo, I.F.; Jung, S.; Gommerman, J.L.; et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 2011, 35, 780–791. [Google Scholar] [CrossRef] [Green Version]
- Mayer, J.U.; Demiri, M.; Agace, W.W.; MacDonald, A.S.; Svensson-Frej, M.; Milling, S.W. Different populations of CD11b(+) dendritic cells drive Th2 responses in the small intestine and colon. Nat. Commun. 2017, 8, 15820. [Google Scholar] [CrossRef]
- Satpathy, A.T.; Briseno, C.G.; Lee, J.S.; Ng, D.; Manieri, N.A.; Kc, W.; Wu, X.; Thomas, S.R.; Lee, W.L.; Turkoz, M.; et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 2013, 14, 937–948. [Google Scholar] [CrossRef]
- Schlitzer, A.; McGovern, N.; Teo, P.; Zelante, T.; Atarashi, K.; Low, D.; Ho, A.W.; See, P.; Shin, A.; Wasan, P.S.; et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 2013, 38, 970–983. [Google Scholar] [CrossRef] [Green Version]
- Tussiwand, R.; Everts, B.; Grajales-Reyes, G.E.; Kretzer, N.M.; Iwata, A.; Bagaitkar, J.; Wu, X.; Wong, R.; Anderson, D.A.; Murphy, T.L.; et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 2015, 42, 916–928. [Google Scholar] [CrossRef] [Green Version]
- Williams, J.W.; Tjota, M.Y.; Clay, B.S.; Vander Lugt, B.; Bandukwala, H.S.; Hrusch, C.L.; Decker, D.C.; Blaine, K.M.; Fixsen, B.R.; Singh, H.; et al. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat. Commun. 2013, 4, 2990. [Google Scholar] [CrossRef] [Green Version]
- Barr, T.A.; Brown, S.; Ryan, G.; Zhao, J.; Gray, D. TLR-mediated stimulation of APC: Distinct cytokine responses of B cells and dendritic cells. Eur. J. Immunol. 2007, 37, 3040–3053. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [Green Version]
- Loschko, J.; Heink, S.; Hackl, D.; Dudziak, D.; Reindl, W.; Korn, T.; Krug, A.B. Antigen targeting to plasmacytoid dendritic cells via Siglec-H inhibits Th cell-dependent autoimmunity. J. Immunol. (Baltim. Md. 1950) 2011, 187, 6346–6356. [Google Scholar] [CrossRef] [PubMed]
- Mouries, J.; Moron, G.; Schlecht, G.; Escriou, N.; Dadaglio, G.; Leclerc, C. Plasmacytoid dendritic cells efficiently cross-prime naive T cells in vivo after TLR activation. Blood 2008, 112, 3713–3722. [Google Scholar] [CrossRef] [PubMed]
- Tel, J.; Aarntzen, E.H.; Baba, T.; Schreibelt, G.; Schulte, B.M.; Benitez-Ribas, D.; Boerman, O.C.; Croockewit, S.; Oyen, W.J.; van Rossum, M.; et al. Natural human plasmacytoid dendritic cells induce antigen-specific T-cell responses in melanoma patients. Cancer Res. 2013, 73, 1063–1075. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Gregorio, J.D.; Iwahori, T.; Zhang, X.; Choi, O.; Tolentino, L.L.; Prestwood, T.; Carmi, Y.; Engleman, E.G. A distinct subset of plasmacytoid dendritic cells induces activation and differentiation of B and T lymphocytes. Proc. Natl. Acad. Sci. USA 2017, 114, 1988–1993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores, M.; Desai, D.D.; Downie, M.; Liang, B.; Reilly, M.P.; McKenzie, S.E.; Clynes, R. Dominant expression of the inhibitory FcgammaRIIB prevents antigen presentation by murine plasmacytoid dendritic cells. J. Immunol. (Baltim. Md. 1950) 2009, 183, 7129–7139. [Google Scholar] [CrossRef] [Green Version]
- Sapoznikov, A.; Fischer, J.A.; Zaft, T.; Krauthgamer, R.; Dzionek, A.; Jung, S. Organ-dependent in vivo priming of naive CD4+, but not CD8+, T cells by plasmacytoid dendritic cells. J. Exp. Med. 2007, 204, 1923–1933. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.C.; Gudjonson, H.; Pritykin, Y.; Deep, D.; Lavallee, V.P.; Mendoza, A.; Fromme, R.; Mazutis, L.; Ariyan, C.; Leslie, C.; et al. Transcriptional Basis of Mouse and Human Dendritic Cell Heterogeneity. Cell 2019, 179, 846–863.e824. [Google Scholar] [CrossRef] [Green Version]
- Swiecki, M.; Wang, Y.; Riboldi, E.; Kim, A.H.; Dzutsev, A.; Gilfillan, S.; Vermi, W.; Ruedl, C.; Trinchieri, G.; Colonna, M. Cell depletion in mice that express diphtheria toxin receptor under the control of SiglecH encompasses more than plasmacytoid dendritic cells. J. Immunol. (Baltim. Md. 1950) 2014, 192, 4409–4416. [Google Scholar] [CrossRef] [Green Version]
- Leylek, R.; Alcantara-Hernandez, M.; Lanzar, Z.; Ludtke, A.; Perez, O.A.; Reizis, B.; Idoyaga, J. Integrated Cross-Species Analysis Identifies a Conserved Transitional Dendritic Cell Population. Cell Rep. 2019, 29, 3736–3750.e3738. [Google Scholar] [CrossRef] [Green Version]
- Dress, R.J.; Dutertre, C.A.; Giladi, A.; Schlitzer, A.; Low, I.; Shadan, N.B.; Tay, A.; Lum, J.; Kairi, M.; Hwang, Y.Y.; et al. Plasmacytoid dendritic cells develop from Ly6D(+) lymphoid progenitors distinct from the myeloid lineage. Nat. Immunol. 2019, 20, 852–864. [Google Scholar] [CrossRef]
- Rodrigues, P.F.; Alberti-Servera, L.; Eremin, A.; Grajales-Reyes, G.E.; Ivanek, R.; Tussiwand, R. Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol. 2018, 19, 711–722. [Google Scholar] [CrossRef]
- Alculumbre, S.G.; Saint-Andre, V.; Di Domizio, J.; Vargas, P.; Sirven, P.; Bost, P.; Maurin, M.; Maiuri, P.; Wery, M.; Roman, M.S.; et al. Diversification of human plasmacytoid predendritic cells in response to a single stimulus. Nat. Immunol. 2018, 19, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Segura, E.; Kapp, E.; Gupta, N.; Wong, J.; Lim, J.; Ji, H.; Heath, W.R.; Simpson, R.; Villadangos, J.A. Differential expression of pathogen-recognition molecules between dendritic cell subsets revealed by plasma membrane proteomic analysis. Mol. Immunol. 2010, 47, 1765–1773. [Google Scholar] [CrossRef] [PubMed]
- Haniffa, M.; Shin, A.; Bigley, V.; McGovern, N.; Teo, P.; See, P.; Wasan, P.S.; Wang, X.N.; Malinarich, F.; Malleret, B.; et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity 2012, 37, 60–73. [Google Scholar] [CrossRef] [Green Version]
- Luber, C.A.; Cox, J.; Lauterbach, H.; Fancke, B.; Selbach, M.; Tschopp, J.; Akira, S.; Wiegand, M.; Hochrein, H.; O’Keeffe, M.; et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 2010, 32, 279–289. [Google Scholar] [CrossRef] [Green Version]
- Maier, B.; Leader, A.M.; Chen, S.T.; Tung, N.; Chang, C.; LeBerichel, J.; Chudnovskiy, A.; Maskey, S.; Walker, L.; Finnigan, J.P.; et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 2020, 580, 257–262. [Google Scholar] [CrossRef]
- Dutertre, C.A.; Becht, E.; Irac, S.E.; Khalilnezhad, A.; Narang, V.; Khalilnezhad, S.; Ng, P.Y.; van den Hoogen, L.L.; Leong, J.Y.; Lee, B.; et al. Single-Cell Analysis of Human Mononuclear Phagocytes Reveals Subset-Defining Markers and Identifies Circulating Inflammatory Dendritic Cells. Immunity 2019, 51, 573–589.e578. [Google Scholar] [CrossRef]
- Yin, X.; Yu, H.; Jin, X.; Li, J.; Guo, H.; Shi, Q.; Yin, Z.; Xu, Y.; Wang, X.; Liu, R.; et al. Human Blood CD1c+ Dendritic Cells Encompass CD5high and CD5low Subsets That Differ Significantly in Phenotype, Gene Expression, and Functions. J. Immunol. (Baltim. Md. 1950) 2017, 198, 1553–1564. [Google Scholar] [CrossRef] [Green Version]
- Worah, K.; Mathan, T.S.M.; Vu Manh, T.P.; Keerthikumar, S.; Schreibelt, G.; Tel, J.; Duiveman-de Boer, T.; Skold, A.E.; van Spriel, A.B.; de Vries, I.J.M.; et al. Proteomics of Human Dendritic Cell Subsets Reveals Subset-Specific Surface Markers and Differential Inflammasome Function. Cell Rep. 2016, 16, 2953–2966. [Google Scholar] [CrossRef] [Green Version]
- Vremec, D.; Pooley, J.; Hochrein, H.; Wu, L.; Shortman, K. CD4 and CD8 expression by dendritic cell subtypes in mouse thymus and spleen. J. Immunol. (Baltim. Md. 1950) 2000, 164, 2978–2986. [Google Scholar] [CrossRef] [Green Version]
- Bigley, V.; McGovern, N.; Milne, P.; Dickinson, R.; Pagan, S.; Cookson, S.; Haniffa, M.; Collin, M. Langerin-expressing dendritic cells in human tissues are related to CD1c+ dendritic cells and distinct from Langerhans cells and CD141high XCR1+ dendritic cells. J. Leukoc. Boil. 2015, 97, 627–634. [Google Scholar] [CrossRef]
- Yarovinsky, F.; Zhang, D.; Andersen, J.F.; Bannenberg, G.L.; Serhan, C.N.; Hayden, M.S.; Hieny, S.; Sutterwala, F.S.; Flavell, R.A.; Ghosh, S.; et al. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science (N. Y.) 2005, 308, 1626–1629. [Google Scholar] [CrossRef] [Green Version]
- Robbins, S.H.; Walzer, T.; Dembele, D.; Thibault, C.; Defays, A.; Bessou, G.; Xu, H.; Vivier, E.; Sellars, M.; Pierre, P.; et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 2008, 9, R17. [Google Scholar] [CrossRef] [PubMed]
- Schulz, O.; Diebold, S.S.; Chen, M.; Naslund, T.I.; Nolte, M.A.; Alexopoulou, L.; Azuma, Y.T.; Flavell, R.A.; Liljestrom, P.; Reis e Sousa, C. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature 2005, 433, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.D.; Diebold, S.S.; Slack, E.M.; Tomizawa, H.; Hemmi, H.; Kaisho, T.; Akira, S.; Reis e Sousa, C. Toll-like receptor expression in murine DC subsets: Lack of TLR7 expression by CD8 alpha+ DC correlates with unresponsiveness to imidazoquinolines. Eur. J. Immunol. 2003, 33, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Lahoud, M.H.; Proietto, A.I.; Ahmet, F.; Kitsoulis, S.; Eidsmo, L.; Wu, L.; Sathe, P.; Pietersz, S.; Chang, H.W.; Walker, I.D.; et al. The C-type lectin Clec12A present on mouse and human dendritic cells can serve as a target for antigen delivery and enhancement of antibody responses. J. Immunol. (Baltim. Md. 1950) 2009, 182, 7587–7594. [Google Scholar] [CrossRef]
- Hutten, T.J.; Thordardottir, S.; Fredrix, H.; Janssen, L.; Woestenenk, R.; Tel, J.; Joosten, B.; Cambi, A.; Heemskerk, M.H.; Franssen, G.M.; et al. CLEC12A-Mediated Antigen Uptake and Cross-Presentation by Human Dendritic Cell Subsets Efficiently Boost Tumor-Reactive T Cell Responses. J. Immunol. (Baltim. Md. 1950) 2016, 197, 2715–2725. [Google Scholar] [CrossRef]
- Flinsenberg, T.W.; Compeer, E.B.; Koning, D.; Klein, M.; Amelung, F.J.; van Baarle, D.; Boelens, J.J.; Boes, M. Fcgamma receptor antigen targeting potentiates cross-presentation by human blood and lymphoid tissue BDCA-3+ dendritic cells. Blood 2012, 120, 5163–5172. [Google Scholar] [CrossRef] [Green Version]
- Guilliams, M.; Bruhns, P.; Saeys, Y.; Hammad, H.; Lambrecht, B.N. The function of Fcgamma receptors in dendritic cells and macrophages. Nat. Rev. Immunol. 2014, 14, 94–108. [Google Scholar] [CrossRef]
- Iberg, C.A.; Jones, A.; Hawiger, D. Dendritic Cells As Inducers of Peripheral Tolerance. Trends Immunol. 2017, 38, 793–804. [Google Scholar] [CrossRef]
- Iwasaki, A.; Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015, 16, 343–353. [Google Scholar] [CrossRef]
- Jain, A.; Pasare, C. Innate Control of Adaptive Immunity: Beyond the Three-Signal Paradigm. J. Immunol. (Baltim. Md. 1950) 2017, 198, 3791–3800. [Google Scholar] [CrossRef] [Green Version]
- Wculek, S.K.; Cueto, F.J.; Mujal, A.M.; Melero, I.; Krummel, M.F.; Sancho, D. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2020, 20, 7–24. [Google Scholar] [CrossRef]
- Broz, P.; Monack, D.M. Newly described pattern recognition receptors team up against intracellular pathogens. Nat. Rev. Immunol. 2013, 13, 551–565. [Google Scholar] [CrossRef]
- Cao, X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 2016, 16, 35–50. [Google Scholar] [CrossRef]
- Lehmann, C.H.; Heger, L.; Heidkamp, G.F.; Baranska, A.; Luhr, J.J.; Hoffmann, A.; Dudziak, D. Direct Delivery of Antigens to Dendritic Cells via Antibodies Specific for Endocytic Receptors as a Promising Strategy for Future Therapies. Vaccines 2016, 4, 8. [Google Scholar] [CrossRef]
- Vatner, R.E.; Janssen, E.M. STING, DCs and the link between innate and adaptive tumor immunity. Mol. Immunol. 2019, 110, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Barber, G.N. STING: Infection, inflammation and cancer. Nat. Rev. Immunol. 2015, 15, 760–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, G.D.; Willment, J.A.; Whitehead, L. C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 2018, 18, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef] [Green Version]
- Gay, N.J.; Symmons, M.F.; Gangloff, M.; Bryant, C.E. Assembly and localization of Toll-like receptor signalling complexes. Nat. Rev. Immunol. 2014, 14, 546–558. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Golenbock, D.; Bowie, A.G. The history of Toll-like receptors—redefining innate immunity. Nat. Rev. Immunol. 2013, 13, 453–460. [Google Scholar] [CrossRef]
- Philpott, D.J.; Sorbara, M.T.; Robertson, S.J.; Croitoru, K.; Girardin, S.E. NOD proteins: Regulators of inflammation in health and disease. Nat. Rev. Immunol. 2014, 14, 9–23. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Tan, C.P.; Goubau, D.; Schulz, O.; Pichlmair, A.; Bier, K.; Robb, N.; Vreede, F.; Barclay, W.; Fodor, E.; et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 2010, 140, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Swanson, K.V.; Deng, M.; Ting, J.P. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Leulier, F.; Lemaitre, B. Toll-like receptors--taking an evolutionary approach. Nat. Rev. Genet. 2008, 9, 165–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Guiducci, C.; Gong, M.; Cepika, A.M.; Xu, Z.; Tripodo, C.; Bennett, L.; Crain, C.; Quartier, P.; Cush, J.J.; Pascual, V.; et al. RNA recognition by human TLR8 can lead to autoimmune inflammation. J. Exp. Med. 2013, 210, 2903–2919. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oldenburg, M.; Kruger, A.; Ferstl, R.; Kaufmann, A.; Nees, G.; Sigmund, A.; Bathke, B.; Lauterbach, H.; Suter, M.; Dreher, S.; et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science (N. Y.) 2012, 337, 1111–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade, W.A.; Souza Mdo, C.; Ramos-Martinez, E.; Nagpal, K.; Dutra, M.S.; Melo, M.B.; Bartholomeu, D.C.; Ghosh, S.; Golenbock, D.T.; Gazzinelli, R.T. Combined action of nucleic acid-sensing Toll-like receptors and TLR11/TLR12 heterodimers imparts resistance to Toxoplasma gondii in mice. Cell Host Microbe 2013, 13, 42–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathur, R.; Oh, H.; Zhang, D.; Park, S.G.; Seo, J.; Koblansky, A.; Hayden, M.S.; Ghosh, S. A mouse model of Salmonella typhi infection. Cell 2012, 151, 590–602. [Google Scholar] [CrossRef] [Green Version]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- Wang, C.; Chen, T.; Zhang, J.; Yang, M.; Li, N.; Xu, X.; Cao, X. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat. Immunol. 2009, 10, 744–752. [Google Scholar] [CrossRef]
- Rathinam, V.A.; Vanaja, S.K.; Waggoner, L.; Sokolovska, A.; Becker, C.; Stuart, L.M.; Leong, J.M.; Fitzgerald, K.A. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell 2012, 150, 606–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sancho, D.; Reis e Sousa, C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu. Rev. Immunol. 2012, 30, 491–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelensky, A.N.; Gready, J.E. The C-type lectin-like domain superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef]
- Figdor, C.G.; van Kooyk, Y.; Adema, G.J. C-type lectin receptors on dendritic cells and Langerhans cells. Nat. Rev. Immunol. 2002, 2, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Amorim, K.N.; Rampazo, E.V.; Antonialli, R.; Yamamoto, M.M.; Rodrigues, M.M.; Soares, I.S.; Boscardin, S.B. The presence of T cell epitopes is important for induction of antibody responses against antigens directed to DEC205(+) dendritic cells. Sci. Rep. 2016, 6, 39250. [Google Scholar] [CrossRef]
- Boscardin, S.B.; Hafalla, J.C.; Masilamani, R.F.; Kamphorst, A.O.; Zebroski, H.A.; Rai, U.; Morrot, A.; Zavala, F.; Steinman, R.M.; Nussenzweig, R.S.; et al. Antigen targeting to dendritic cells elicits long-lived T cell help for antibody responses. J. Exp. Med. 2006, 203, 599–606. [Google Scholar] [CrossRef]
- Do, Y.; Koh, H.; Park, C.G.; Dudziak, D.; Seo, P.; Mehandru, S.; Choi, J.H.; Cheong, C.; Park, S.; Perlin, D.S.; et al. Targeting of LcrV virulence protein from Yersinia pestis to dendritic cells protects mice against pneumonic plague. Eur. J. Immunol. 2010, 40, 2791–2796. [Google Scholar] [CrossRef]
- Heidkamp, G.F.; Neubert, K.; Haertel, E.; Nimmerjahn, F.; Nussenzweig, M.C.; Dudziak, D. Efficient generation of a monoclonal antibody against the human C-type lectin receptor DCIR by targeting murine dendritic cells. Immunol. Lett. 2010, 132, 69–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahoud, M.H.; Ahmet, F.; Kitsoulis, S.; Wan, S.S.; Vremec, D.; Lee, C.N.; Phipson, B.; Shi, W.; Smyth, G.K.; Lew, A.M.; et al. Targeting antigen to mouse dendritic cells via Clec9A induces potent CD4 T cell responses biased toward a follicular helper phenotype. J. Immunol. (Baltim. Md. 1950) 2011, 187, 842–850. [Google Scholar] [CrossRef] [Green Version]
- Neubert, K.; Lehmann, C.H.; Heger, L.; Baranska, A.; Staedtler, A.M.; Buchholz, V.R.; Yamazaki, S.; Heidkamp, G.F.; Eissing, N.; Zebroski, H.; et al. Antigen delivery to CD11c+CD8- dendritic cells induces protective immune responses against experimental melanoma in mice in vivo. J. Immunol. (Baltim. Md. 1950) 2014, 192, 5830–5838. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Zaidi, N.; He, L.Z.; Zhang, L.; Kuroiwa, J.M.; Keler, T.; Steinman, R.M. Targeting of the non-mutated tumor antigen HER2/neu to mature dendritic cells induces an integrated immune response that protects against breast cancer in mice. Breast Cancer Res. BCR 2012, 14, R39. [Google Scholar] [CrossRef]
- Zaneti, A.B.; Yamamoto, M.M.; Sulczewski, F.B.; Almeida, B.D.S.; Souza, H.F.S.; Ferreira, N.S.; Maeda, D.; Sales, N.S.; Rosa, D.S.; Ferreira, L.C.S.; et al. Dendritic Cell Targeting Using a DNA Vaccine Induces Specific Antibodies and CD4(+) T Cells to the Dengue Virus Envelope Protein Domain III. Front. Immunol. 2019, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, N.; Tashiro, K.; Inaba, K.; Miyachi, Y. Dendritic cell immunoactivating receptor, a novel C-type lectin immunoreceptor, acts as an activating receptor through association with Fc receptor gamma chain. J. Boil. Chem. 2003, 278, 32645–32652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, K.; Yang, X.L.; Yudate, T.; Chung, J.S.; Wu, J.; Luby-Phelps, K.; Kimberly, R.P.; Underhill, D.; Cruz, P.D., Jr.; Ariizumi, K. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J. Boil. Chem. 2006, 281, 38854–38866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamasaki, S.; Ishikawa, E.; Sakuma, M.; Hara, H.; Ogata, K.; Saito, T. Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat. Immunol. 2008, 9, 1179–1188. [Google Scholar] [CrossRef]
- Ahrens, S.; Zelenay, S.; Sancho, D.; Hanc, P.; Kjaer, S.; Feest, C.; Fletcher, G.; Durkin, C.; Postigo, A.; Skehel, M.; et al. F-actin is an evolutionarily conserved damage-associated molecular pattern recognized by DNGR-1, a receptor for dead cells. Immunity 2012, 36, 635–645. [Google Scholar] [CrossRef] [Green Version]
- Cambi, A.; Figdor, C. Necrosis: C-type lectins sense cell death. Curr. Boil. CB 2009, 19, R375–R378. [Google Scholar] [CrossRef] [Green Version]
- Shrimpton, R.E.; Butler, M.; Morel, A.S.; Eren, E.; Hue, S.S.; Ritter, M.A. CD205 (DEC-205): A recognition receptor for apoptotic and necrotic self. Mol. Immunol. 2009, 46, 1229–1239. [Google Scholar] [CrossRef]
- Schreibelt, G.; Klinkenberg, L.J.; Cruz, L.J.; Tacken, P.J.; Tel, J.; Kreutz, M.; Adema, G.J.; Brown, G.D.; Figdor, C.G.; de Vries, I.J. The C-type lectin receptor CLEC9A mediates antigen uptake and (cross-)presentation by human blood BDCA3+ myeloid dendritic cells. Blood 2012, 119, 2284–2292. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, L.X.; Pang, P.; Cui, Z.; Aung, S.; Haley, D.; Fox, B.A.; Urba, W.J.; Hu, H.M. Tumor-derived autophagosome vaccine: Mechanism of cross-presentation and therapeutic efficacy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 7047–7057. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Cao, R.; Hu, H.M. TLR and NLRP3 inflammasome-dependent innate immune responses to tumor-derived autophagosomes (DRibbles). Cell Death Dis. 2016, 7, e2322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, W.; Xing, Y.; Paustian, C.; van de Ven, R.; Moudgil, T.; Hilton, T.L.; Fox, B.A.; Urba, W.J.; Zhao, W.; Hu, H.M. Cross-presentation of viral antigens in dribbles leads to efficient activation of virus-specific human memory T cells. J. Transl. Med. 2014, 12, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yewdell, J.W.; Anton, L.C.; Bennink, J.R. Defective ribosomal products (DRiPs): A major source of antigenic peptides for MHC class I molecules? J. Immunol. (Baltim. Md. 1950) 1996, 157, 1823–1826. [Google Scholar]
- Bournazos, S.; Wang, T.T.; Dahan, R.; Maamary, J.; Ravetch, J.V. Signaling by Antibodies: Recent Progress. Annu. Rev. Immunol. 2017, 35, 285–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nimmerjahn, F.; Gordan, S.; Lux, A. FcgammaR dependent mechanisms of cytotoxic, agonistic, and neutralizing antibody activities. Trends Immunol. 2015, 36, 325–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nimmerjahn, F.; Ravetch, J.V. Fcgamma receptors: Old friends and new family members. Immunity 2006, 24, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Nimmerjahn, F.; Ravetch, J.V. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 2008, 8, 34–47. [Google Scholar] [CrossRef]
- Loo, Y.M.; Gale, M., Jr. Immune signaling by RIG-I-like receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, K.R.; Bruns, A.M.; Horvath, C.M. MDA5 and LGP2: Accomplices and antagonists of antiviral signal transduction. J. Virol. 2014, 88, 8194–8200. [Google Scholar] [CrossRef] [Green Version]
- Sanchez David, R.Y.; Combredet, C.; Najburg, V.; Millot, G.A.; Beauclair, G.; Schwikowski, B.; Leger, T.; Camadro, J.M.; Jacob, Y.; Bellalou, J.; et al. LGP2 binds to PACT to regulate RIG-I- and MDA5-mediated antiviral responses. Sci. Signal. 2019, 12, eaar3993. [Google Scholar] [CrossRef]
- Ablasser, A.; Bauernfeind, F.; Hartmann, G.; Latz, E.; Fitzgerald, K.A.; Hornung, V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 2009, 10, 1065–1072. [Google Scholar] [CrossRef] [Green Version]
- Wilson, N.S.; Behrens, G.M.; Lundie, R.J.; Smith, C.M.; Waithman, J.; Young, L.; Forehan, S.P.; Mount, A.; Steptoe, R.J.; Shortman, K.D.; et al. Systemic activation of dendritic cells by Toll-like receptor ligands or malaria infection impairs cross-presentation and antiviral immunity. Nat. Immunol. 2006, 7, 165–172. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Pantel, A.; Teixeira, A.; Haddad, E.; Wood, E.G.; Steinman, R.M.; Longhi, M.P. Direct type I IFN but not MDA5/TLR3 activation of dendritic cells is required for maturation and metabolic shift to glycolysis after poly IC stimulation. PLoS Biol. 2014, 12, e1001759. [Google Scholar] [CrossRef]
- Desch, A.N.; Gibbings, S.L.; Clambey, E.T.; Janssen, W.J.; Slansky, J.E.; Kedl, R.M.; Henson, P.M.; Jakubzick, C. Dendritic cell subsets require cis-activation for cytotoxic CD8 T-cell induction. Nat. Commun. 2014, 5, 4674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harton, J.A.; Linhoff, M.W.; Zhang, J.; Ting, J.P. Cutting edge: CATERPILLER: A large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J. Immunol. (Baltim. Md. 1950) 2002, 169, 4088–4093. [Google Scholar] [CrossRef] [Green Version]
- Velloso, F.J.; Trombetta-Lima, M.; Anschau, V.; Sogayar, M.C.; Correa, R.G. NOD-like receptors: Major players (and targets) in the interface between innate immunity and cancer. Biosci. Rep. 2019, 39, BSR20181709. [Google Scholar] [CrossRef] [Green Version]
- Warner, N.; Burberry, A.; Franchi, L.; Kim, Y.G.; McDonald, C.; Sartor, M.A.; Nunez, G. A genome-wide siRNA screen reveals positive and negative regulators of the NOD2 and NF-kappaB signaling pathways. Sci. Signal. 2013, 6, rs3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeretssian, G.; Correa, R.G.; Doiron, K.; Fitzgerald, P.; Dillon, C.P.; Green, D.R.; Reed, J.C.; Saleh, M. Non-apoptotic role of BID in inflammation and innate immunity. Nature 2011, 474, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, J.G.; Fritz, J.H.; Le Bourhis, L.; Sellge, G.; Travassos, L.H.; Selvanantham, T.; Girardin, S.E.; Gommerman, J.L.; Philpott, D.J. Nod2-dependent Th2 polarization of antigen-specific immunity. J. Immunol. (Baltim. Md. 1950) 2008, 181, 7925–7935. [Google Scholar] [CrossRef] [Green Version]
- Magalhaes, J.G.; Rubino, S.J.; Travassos, L.H.; Le Bourhis, L.; Duan, W.; Sellge, G.; Geddes, K.; Reardon, C.; Lechmann, M.; Carneiro, L.A.; et al. Nucleotide oligomerization domain-containing proteins instruct T cell helper type 2 immunity through stromal activation. Proc. Natl. Acad. Sci. USA 2011, 108, 14896–14901. [Google Scholar] [CrossRef] [Green Version]
- Fritz, J.H.; Le Bourhis, L.; Sellge, G.; Magalhaes, J.G.; Fsihi, H.; Kufer, T.A.; Collins, C.; Viala, J.; Ferrero, R.L.; Girardin, S.E.; et al. Nod1-mediated innate immune recognition of peptidoglycan contributes to the onset of adaptive immunity. Immunity 2007, 26, 445–459. [Google Scholar] [CrossRef] [Green Version]
- Cooney, R.; Baker, J.; Brain, O.; Danis, B.; Pichulik, T.; Allan, P.; Ferguson, D.J.; Campbell, B.J.; Jewell, D.; Simmons, A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 2010, 16, 90–97. [Google Scholar] [CrossRef]
- Homer, C.R.; Kabi, A.; Marina-Garcia, N.; Sreekumar, A.; Nesvizhskii, A.I.; Nickerson, K.P.; Chinnaiyan, A.M.; Nunez, G.; McDonald, C. A dual role for receptor-interacting protein kinase 2 (RIP2) kinase activity in nucleotide-binding oligomerization domain 2 (NOD2)-dependent autophagy. J. Boil. Chem. 2012, 287, 25565–25576. [Google Scholar] [CrossRef] [Green Version]
- Homer, C.R.; Richmond, A.L.; Rebert, N.A.; Achkar, J.P.; McDonald, C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology 2010, 139, 1630–1641, e1631–e1632. [Google Scholar] [CrossRef] [Green Version]
- Lupfer, C.; Thomas, P.G.; Anand, P.K.; Vogel, P.; Milasta, S.; Martinez, J.; Huang, G.; Green, M.; Kundu, M.; Chi, H.; et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 2013, 14, 480–488. [Google Scholar] [CrossRef] [Green Version]
- Travassos, L.H.; Carneiro, L.A.; Ramjeet, M.; Hussey, S.; Kim, Y.G.; Magalhaes, J.G.; Yuan, L.; Soares, F.; Chea, E.; Le Bourhis, L.; et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010, 11, 55–62. [Google Scholar] [CrossRef]
- Wen, H.; Miao, E.A.; Ting, J.P. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity 2013, 39, 432–441. [Google Scholar] [CrossRef] [Green Version]
- Bauernfeind, F.G.; Horvath, G.; Stutz, A.; Alnemri, E.S.; MacDonald, K.; Speert, D.; Fernandes-Alnemri, T.; Wu, J.; Monks, B.G.; Fitzgerald, K.A.; et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. (Baltim. Md. 1950) 2009, 183, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Franchi, L.; Eigenbrod, T.; Nunez, G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. (Baltim. Md. 1950) 2009, 183, 792–796. [Google Scholar] [CrossRef]
- Xing, Y.; Yao, X.; Li, H.; Xue, G.; Guo, Q.; Yang, G.; An, L.; Zhang, Y.; Meng, G. Cutting Edge: TRAF6 Mediates TLR/IL-1R Signaling-Induced Nontranscriptional Priming of the NLRP3 Inflammasome. J. Immunol. (Baltim. Md. 1950) 2017, 199, 1561–1566. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Chen, J.; Xu, H.; Liu, S.; Jiang, Q.X.; Halfmann, R.; Chen, Z.J. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 2014, 156, 1207–1222. [Google Scholar] [CrossRef] [Green Version]
- Lu, A.; Magupalli, V.G.; Ruan, J.; Yin, Q.; Atianand, M.K.; Vos, M.R.; Schroder, G.F.; Fitzgerald, K.A.; Wu, H.; Egelman, E.H. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 2014, 156, 1193–1206. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, F.I.; Lu, A.; Chen, J.W.; Ruan, J.; Tang, C.; Wu, H.; Ploegh, H.L. A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly. J. Exp. Med. 2016, 213, 771–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Zeng, M.Y.; Yang, D.; Motro, B.; Nunez, G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 2016, 530, 354–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Wang, Y.; Li, X.; Zhan, X.; Tang, M.; Fina, M.; Su, L.; Pratt, D.; Bu, C.H.; Hildebrand, S.; et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 2016, 17, 250–258. [Google Scholar] [CrossRef]
- He, Y.; Franchi, L.; Nunez, G. TLR agonists stimulate Nlrp3-dependent IL-1beta production independently of the purinergic P2X7 receptor in dendritic cells and in vivo. J. Immunol. (Baltim. Md. 1950) 2013, 190, 334–339. [Google Scholar] [CrossRef] [Green Version]
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O’Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T.; et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.L.; Stowe, I.B.; Gupta, A.; Kornfeld, O.S.; Roose-Girma, M.; Anderson, K.; Warming, S.; Zhang, J.; Lee, W.P.; Kayagaki, N. Caspase-11 auto-proteolysis is crucial for noncanonical inflammasome activation. J. Exp. Med. 2018, 215, 2279–2288. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Wang, K.; Liu, W.; She, Y.; Sun, Q.; Shi, J.; Sun, H.; Wang, D.C.; Shao, F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016, 535, 111–116. [Google Scholar] [CrossRef]
- He, W.T.; Wan, H.; Hu, L.; Chen, P.; Wang, X.; Huang, Z.; Yang, Z.H.; Zhong, C.Q.; Han, J. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 2015, 25, 1285–1298. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Z.; Ruan, J.; Pan, Y.; Magupalli, V.G.; Wu, H.; Lieberman, J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016, 535, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Zanoni, I.; Tan, Y.; Di Gioia, M.; Broggi, A.; Ruan, J.; Shi, J.; Donado, C.A.; Shao, F.; Wu, H.; Springstead, J.R.; et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science (N. Y.) 2016, 352, 1232–1236. [Google Scholar] [CrossRef] [Green Version]
- Monteleone, M.; Stanley, A.C.; Chen, K.W.; Brown, D.L.; Bezbradica, J.S.; von Pein, J.B.; Holley, C.L.; Boucher, D.; Shakespear, M.R.; Kapetanovic, R.; et al. Interleukin-1beta Maturation Triggers Its Relocation to the Plasma Membrane for Gasdermin-D-Dependent and -Independent Secretion. Cell Rep. 2018, 24, 1425–1433. [Google Scholar] [CrossRef] [Green Version]
- Zanoni, I.; Tan, Y.; Di Gioia, M.; Springstead, J.R.; Kagan, J.C. By Capturing Inflammatory Lipids Released from Dying Cells, the Receptor CD14 Induces Inflammasome-Dependent Phagocyte Hyperactivation. Immunity 2017, 47, 697–709.e693. [Google Scholar] [CrossRef] [Green Version]
- Ghiringhelli, F.; Apetoh, L.; Tesniere, A.; Aymeric, L.; Ma, Y.; Ortiz, C.; Vermaelen, K.; Panaretakis, T.; Mignot, G.; Ullrich, E.; et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat. Med. 2009, 15, 1170–1178. [Google Scholar] [CrossRef]
- Lee, Y.K.; Turner, H.; Maynard, C.L.; Oliver, J.R.; Chen, D.; Elson, C.O.; Weaver, C.T. Late developmental plasticity in the T helper 17 lineage. Immunity 2009, 30, 92–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Godec, J.; Ben-Aissa, K.; Cui, K.; Zhao, K.; Pucsek, A.B.; Lee, Y.K.; Weaver, C.T.; Yagi, R.; Lazarevic, V. The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-gamma-producing T helper 17 cells. Immunity 2014, 40, 355–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, G.; Wei, L.; Zhu, J.; Zang, C.; Hu-Li, J.; Yao, Z.; Cui, K.; Kanno, Y.; Roh, T.Y.; Watford, W.T.; et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 2009, 30, 155–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zielinski, C.E.; Mele, F.; Aschenbrenner, D.; Jarrossay, D.; Ronchi, F.; Gattorno, M.; Monticelli, S.; Lanzavecchia, A.; Sallusto, F. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature 2012, 484, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef]
- Christgen, S.; Place, D.E.; Kanneganti, T.D. Toward targeting inflammasomes: Insights into their regulation and activation. Cell Res. 2020, 30, 315–327. [Google Scholar] [CrossRef] [Green Version]
- Burdette, D.L.; Monroe, K.M.; Sotelo-Troha, K.; Iwig, J.S.; Eckert, B.; Hyodo, M.; Hayakawa, Y.; Vance, R.E. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011, 478, 515–518. [Google Scholar] [CrossRef]
- Burdette, D.L.; Vance, R.E. STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 2013, 14, 19–26. [Google Scholar] [CrossRef]
- Ishikawa, H.; Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008, 455, 674–678. [Google Scholar] [CrossRef]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef] [Green Version]
- Sauer, J.D.; Sotelo-Troha, K.; von Moltke, J.; Monroe, K.M.; Rae, C.S.; Brubaker, S.W.; Hyodo, M.; Hayakawa, Y.; Woodward, J.J.; Portnoy, D.A.; et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 2011, 79, 688–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science (N. Y.) 2013, 339, 786–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Li, Y.; Chen, L.; Chen, H.; You, F.; Zhou, X.; Zhou, Y.; Zhai, Z.; Chen, D.; Jiang, Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA 2009, 106, 8653–8658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodward, J.J.; Iavarone, A.T.; Portnoy, D.A. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science (N. Y.) 2010, 328, 1703–1705. [Google Scholar] [CrossRef] [Green Version]
- Zhong, B.; Yang, Y.; Li, S.; Wang, Y.Y.; Li, Y.; Diao, F.; Lei, C.; He, X.; Zhang, L.; Tien, P.; et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 2008, 29, 538–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ablasser, A.; Schmid-Burgk, J.L.; Hemmerling, I.; Horvath, G.L.; Schmidt, T.; Latz, E.; Hornung, V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 2013, 503, 530–534. [Google Scholar] [CrossRef] [Green Version]
- Diner, E.J.; Burdette, D.L.; Wilson, S.C.; Monroe, K.M.; Kellenberger, C.A.; Hyodo, M.; Hayakawa, Y.; Hammond, M.C.; Vance, R.E. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013, 3, 1355–1361. [Google Scholar] [CrossRef] [Green Version]
- McWhirter, S.M.; Barbalat, R.; Monroe, K.M.; Fontana, M.F.; Hyodo, M.; Joncker, N.T.; Ishii, K.J.; Akira, S.; Colonna, M.; Chen, Z.J.; et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J. Exp. Med. 2009, 206, 1899–1911. [Google Scholar] [CrossRef]
- Salcedo, R.; Cataisson, C.; Hasan, U.; Yuspa, S.H.; Trinchieri, G. MyD88 and its divergent toll in carcinogenesis. Trends Immunol. 2013, 34, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Swann, J.B.; Vesely, M.D.; Silva, A.; Sharkey, J.; Akira, S.; Schreiber, R.D.; Smyth, M.J. Demonstration of inflammation-induced cancer and cancer immunoediting during primary tumorigenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 652–656. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.; Xia, T.; Konno, H.; Konno, K.; Ruiz, P.; Barber, G.N. Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 2014, 5, 5166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, J.; Konno, H.; Barber, G.N. Diverse roles of STING-dependent signaling on the development of cancer. Oncogene 2015, 34, 5302–5308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Li, L.; Lemos, H.; Chandler, P.R.; Pacholczyk, G.; Baban, B.; Barber, G.N.; Hayakawa, Y.; McGaha, T.L.; Ravishankar, B.; et al. Cutting edge: DNA sensing via the STING adaptor in myeloid dendritic cells induces potent tolerogenic responses. J. Immunol. (Baltim. Md. 1950) 2013, 191, 3509–3513. [Google Scholar] [CrossRef] [Green Version]
- Lemos, H.; Mohamed, E.; Huang, L.; Ou, R.; Pacholczyk, G.; Arbab, A.S.; Munn, D.; Mellor, A.L. STING Promotes the Growth of Tumors Characterized by Low Antigenicity via IDO Activation. Cancer Res. 2016, 76, 2076–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Q.; Man, S.M.; Gurung, P.; Liu, Z.; Vogel, P.; Lamkanfi, M.; Kanneganti, T.D. Cutting edge: STING mediates protection against colorectal tumorigenesis by governing the magnitude of intestinal inflammation. J. Immunol. (Baltim. Md. 1950) 2014, 193, 4779–4782. [Google Scholar] [CrossRef]
- Marcus, A.; Mao, A.J.; Lensink-Vasan, M.; Wang, L.; Vance, R.E.; Raulet, D.H. Tumor-Derived cGAMP Triggers a STING-Mediated Interferon Response in Non-tumor Cells to Activate the NK Cell Response. Immunity 2018, 49, 754–763.e754. [Google Scholar] [CrossRef] [Green Version]
- Hartlova, A.; Erttmann, S.F.; Raffi, F.A.; Schmalz, A.M.; Resch, U.; Anugula, S.; Lienenklaus, S.; Nilsson, L.M.; Kroger, A.; Nilsson, J.A.; et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 2015, 42, 332–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, T.; Kobayashi, J.; Saitoh, T.; Maruyama, K.; Ishii, K.J.; Barber, G.N.; Komatsu, K.; Akira, S.; Kawai, T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl. Acad. Sci. USA 2013, 110, 2969–2974. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Kanne, D.B.; Leong, M.; Glickman, L.H.; McWhirter, S.M.; Lemmens, E.; Mechette, K.; Leong, J.J.; Lauer, P.; Liu, W.; et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med. 2015, 7, 283ra252. [Google Scholar] [CrossRef] [Green Version]
- Ohkuri, T.; Ghosh, A.; Kosaka, A.; Zhu, J.; Ikeura, M.; David, M.; Watkins, S.C.; Sarkar, S.N.; Okada, H. STING contributes to antiglioma immunity via triggering type I IFN signals in the tumor microenvironment. Cancer Immunol. Res. 2014, 2, 1199–1208. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.R.; Fuertes, M.B.; Corrales, L.; Spranger, S.; Furdyna, M.J.; Leung, M.Y.; Duggan, R.; Wang, Y.; Barber, G.N.; Fitzgerald, K.A.; et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 2014, 41, 830–842. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.; Gutman, D.; Saijo, S.; Barber, G.N. STING manifests self DNA-dependent inflammatory disease. Proc. Natl. Acad. Sci. USA 2012, 109, 19386–19391. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Liang, H.; Xu, M.; Yang, X.; Burnette, B.; Arina, A.; Li, X.D.; Mauceri, H.; Beckett, M.; Darga, T.; et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity 2014, 41, 843–852. [Google Scholar] [CrossRef] [Green Version]
- Klarquist, J.; Hennies, C.M.; Lehn, M.A.; Reboulet, R.A.; Feau, S.; Janssen, E.M. STING-mediated DNA sensing promotes antitumor and autoimmune responses to dying cells. J. Immunol. (Baltim. Md. 1950) 2014, 193, 6124–6134. [Google Scholar] [CrossRef]
- Gonugunta, V.K.; Sakai, T.; Pokatayev, V.; Yang, K.; Wu, J.; Dobbs, N.; Yan, N. Trafficking-Mediated STING Degradation Requires Sorting to Acidified Endolysosomes and Can Be Targeted to Enhance Anti-tumor Response. Cell Rep. 2017, 21, 3234–3242. [Google Scholar] [CrossRef] [Green Version]
- White, M.J.; McArthur, K.; Metcalf, D.; Lane, R.M.; Cambier, J.C.; Herold, M.J.; van Delft, M.F.; Bedoui, S.; Lessene, G.; Ritchie, M.E.; et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 2014, 159, 1549–1562. [Google Scholar] [CrossRef] [Green Version]
- Corrales, L.; Gajewski, T.F. Molecular Pathways: Targeting the Stimulator of Interferon Genes (STING) in the Immunotherapy of Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 4774–4779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrales, L.; Glickman, L.H.; McWhirter, S.M.; Kanne, D.B.; Sivick, K.E.; Katibah, G.E.; Woo, S.R.; Lemmens, E.; Banda, T.; Leong, J.J.; et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep. 2015, 11, 1018–1030. [Google Scholar] [CrossRef] [Green Version]
- Dubensky, T.W., Jr.; Kanne, D.B.; Leong, M.L. Rationale, progress and development of vaccines utilizing STING-activating cyclic dinucleotide adjuvants. Ther. Adv. Vaccines 2013, 1, 131–143. [Google Scholar] [CrossRef]
- Wang, Z.; Celis, E. STING activator c-di-GMP enhances the anti-tumor effects of peptide vaccines in melanoma-bearing mice. Cancer Immunol. Immunother. 2015, 64, 1057–1066. [Google Scholar] [CrossRef]
- Lan, Y.Y.; Londono, D.; Bouley, R.; Rooney, M.S.; Hacohen, N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep. 2014, 9, 180–192. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Crowe, W.N.; Wang, L.; Lu, Y.; Petty, W.J.; Habib, A.A.; Zhao, D. An inhalable nanoparticulate STING agonist synergizes with radiotherapy to confer long-term control of lung metastases. Nat. Commun. 2019, 10, 5108. [Google Scholar] [CrossRef] [Green Version]
- Lohard, S.; Bourgeois, N.; Maillet, L.; Gautier, F.; Fetiveau, A.; Lasla, H.; Nguyen, F.; Vuillier, C.; Dumont, A.; Moreau-Aubry, A.; et al. STING-dependent paracriny shapes apoptotic priming of breast tumors in response to anti-mitotic treatment. Nat. Commun. 2020, 11, 259. [Google Scholar] [CrossRef] [Green Version]
- Zitvogel, L.; Galluzzi, L.; Smyth, M.J.; Kroemer, G. Mechanism of action of conventional and targeted anticancer therapies: Reinstating immunosurveillance. Immunity 2013, 39, 74–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torralba, D.; Baixauli, F.; Villarroya-Beltri, C.; Fernandez-Delgado, I.; Latorre-Pellicer, A.; Acin-Perez, R.; Martin-Cofreces, N.B.; Jaso-Tamame, A.L.; Iborra, S.; Jorge, I.; et al. Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat. Commun. 2018, 9, 2658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, J.; Barber, G.N. STING signaling and host defense against microbial infection. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Li, T.; Chen, Z.J. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 2018, 215, 1287–1299. [Google Scholar] [CrossRef]
- Motwani, M.; Pesiridis, S.; Fitzgerald, K.A. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 2019, 20, 657–674. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.R.; Corrales, L.; Gajewski, T.F. The STING pathway and the T cell-inflamed tumor microenvironment. Trends immunol. 2015, 36, 250–256. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, O.; Sato, S.; Horiuchi, T.; Hoshino, K.; Takeda, K.; Dong, Z.; Modlin, R.L.; Akira, S. Cutting edge: Role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. (Baltim. Md. 1950) 2002, 169, 10–14. [Google Scholar] [CrossRef]
- Funderburg, N.T.; Jadlowsky, J.K.; Lederman, M.M.; Feng, Z.; Weinberg, A.; Sieg, S.F. The Toll-like receptor 1/2 agonists Pam(3) CSK(4) and human beta-defensin-3 differentially induce interleukin-10 and nuclear factor-kappaB signalling patterns in human monocytes. Immunology 2011, 134, 151–160. [Google Scholar] [CrossRef]
- Campos, M.A.; Almeida, I.C.; Takeuchi, O.; Akira, S.; Valente, E.P.; Procopio, D.O.; Travassos, L.R.; Smith, J.A.; Golenbock, D.T.; Gazzinelli, R.T. Activation of Toll-like receptor-2 by glycosylphosphatidylinositol anchors from a protozoan parasite. J. Immunol. (Baltim. Md. 1950) 2001, 167, 416–423. [Google Scholar] [CrossRef] [Green Version]
- Lien, E.; Sellati, T.J.; Yoshimura, A.; Flo, T.H.; Rawadi, G.; Finberg, R.W.; Carroll, J.D.; Espevik, T.; Ingalls, R.R.; Radolf, J.D.; et al. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J. Boil. Chem. 1999, 274, 33419–33425. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef]
- Means, T.K.; Wang, S.; Lien, E.; Yoshimura, A.; Golenbock, D.T.; Fenton, M.J. Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J. Immunol. (Baltim. Md. 1950) 1999, 163, 3920–3927. [Google Scholar]
- Brightbill, H.D.; Libraty, D.H.; Krutzik, S.R.; Yang, R.B.; Belisle, J.T.; Bleharski, J.R.; Maitland, M.; Norgard, M.V.; Plevy, S.E.; Smale, S.T.; et al. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science (N. Y.) 1999, 285, 732–736. [Google Scholar] [CrossRef] [Green Version]
- Aliprantis, A.O.; Yang, R.B.; Mark, M.R.; Suggett, S.; Devaux, B.; Radolf, J.D.; Klimpel, G.R.; Godowski, P.; Zychlinsky, A. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science (N. Y.) 1999, 285, 736–739. [Google Scholar] [CrossRef]
- Yang, R.B.; Mark, M.R.; Gray, A.; Huang, A.; Xie, M.H.; Zhang, M.; Goddard, A.; Wood, W.I.; Gurney, A.L.; Godowski, P.J. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature 1998, 395, 284–288. [Google Scholar] [CrossRef]
- Kirschning, C.J.; Wesche, H.; Merrill Ayres, T.; Rothe, M. Human toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J. Exp. Med. 1998, 188, 2091–2097. [Google Scholar] [CrossRef] [Green Version]
- Schwandner, R.; Dziarski, R.; Wesche, H.; Rothe, M.; Kirschning, C.J. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J. Boil. Chem. 1999, 274, 17406–17409. [Google Scholar] [CrossRef] [Green Version]
- Underhill, D.M.; Ozinsky, A.; Hajjar, A.M.; Stevens, A.; Wilson, C.B.; Bassetti, M.; Aderem, A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature 1999, 401, 811–815. [Google Scholar] [CrossRef]
- Takeuchi, O.; Hoshino, K.; Kawai, T.; Sanjo, H.; Takada, H.; Ogawa, T.; Takeda, K.; Akira, S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 1999, 11, 443–451. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, O.; Kawai, T.; Muhlradt, P.F.; Morr, M.; Radolf, J.D.; Zychlinsky, A.; Takeda, K.; Akira, S. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 2001, 13, 933–940. [Google Scholar] [CrossRef]
- Gnjatic, S.; Sawhney, N.B.; Bhardwaj, N. Toll-like receptor agonists: Are they good adjuvants? Cancer J. 2010, 16, 382–391. [Google Scholar] [CrossRef] [Green Version]
- Vabulas, R.M.; Ahmad-Nejad, P.; Ghose, S.; Kirschning, C.J.; Issels, R.D.; Wagner, H. HSP70 as endogenous stimulus of the Toll/interleukin-1 receptor signal pathway. J. Boil. Chem. 2002, 277, 15107–15112. [Google Scholar] [CrossRef] [Green Version]
- Vabulas, R.M.; Braedel, S.; Hilf, N.; Singh-Jasuja, H.; Herter, S.; Ahmad-Nejad, P.; Kirschning, C.J.; Da Costa, C.; Rammensee, H.G.; Wagner, H.; et al. The endoplasmic reticulum-resident heat shock protein Gp96 activates dendritic cells via the Toll-like receptor 2/4 pathway. J. Boil. Chem. 2002, 277, 20847–20853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vabulas, R.M.; Ahmad-Nejad, P.; da Costa, C.; Miethke, T.; Kirschning, C.J.; Hacker, H.; Wagner, H. Endocytosed HSP60s use toll-like receptor 2 (TLR2) and TLR4 to activate the toll/interleukin-1 receptor signaling pathway in innate immune cells. J. Boil. Chem. 2001, 276, 31332–31339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, D.; Liang, J.; Fan, J.; Yu, S.; Chen, S.; Luo, Y.; Prestwich, G.D.; Mascarenhas, M.M.; Garg, H.G.; Quinn, D.A.; et al. Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat. Med. 2005, 11, 1173–1179. [Google Scholar] [CrossRef]
- Asea, A.; Rehli, M.; Kabingu, E.; Boch, J.A.; Bare, O.; Auron, P.E.; Stevenson, M.A.; Calderwood, S.K. Novel signal transduction pathway utilized by extracellular HSP70: Role of toll-like receptor (TLR) 2 and TLR4. J. Boil. Chem. 2002, 277, 15028–15034. [Google Scholar] [CrossRef] [Green Version]
- Liu-Bryan, R.; Scott, P.; Sydlaske, A.; Rose, D.M.; Terkeltaub, R. Innate immunity conferred by Toll-like receptors 2 and 4 and myeloid differentiation factor 88 expression is pivotal to monosodium urate monohydrate crystal-induced inflammation. Arthritis Rheum 2005, 52, 2936–2946. [Google Scholar] [CrossRef]
- Toussi, D.N.; Massari, P. Immune Adjuvant Effect of Molecularly-defined Toll-Like Receptor Ligands. Vaccines 2014, 2, 323–353. [Google Scholar] [CrossRef]
- Muhlradt, P.F.; Kiess, M.; Meyer, H.; Sussmuth, R.; Jung, G. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J. Exp. Med. 1997, 185, 1951–1958. [Google Scholar] [CrossRef] [Green Version]
- Cataldi, A.; Yevsa, T.; Vilte, D.A.; Schulze, K.; Castro-Parodi, M.; Larzabal, M.; Ibarra, C.; Mercado, E.C.; Guzman, C.A. Efficient immune responses against Intimin and EspB of enterohaemorragic Escherichia coli after intranasal vaccination using the TLR2/6 agonist MALP-2 as adjuvant. Vaccine 2008, 26, 5662–5667. [Google Scholar] [CrossRef]
- Oosenbrug, T.; van de Graaff, M.J.; Ressing, M.E.; van Kasteren, S.I. Chemical Tools for Studying TLR Signaling Dynamics. Cell Chem. Biol. 2017, 24, 801–812. [Google Scholar] [CrossRef] [Green Version]
- Sivori, S.; Falco, M.; Della Chiesa, M.; Carlomagno, S.; Vitale, M.; Moretta, L.; Moretta, A. CpG and double-stranded RNA trigger human NK cells by Toll-like receptors: Induction of cytokine release and cytotoxicity against tumors and dendritic cells. Proc. Natl. Acad. Sci. USA 2004, 101, 10116–10121. [Google Scholar] [CrossRef] [Green Version]
- Cavassani, K.A.; Ishii, M.; Wen, H.; Schaller, M.A.; Lincoln, P.M.; Lukacs, N.W.; Hogaboam, C.M.; Kunkel, S.L. TLR3 is an endogenous sensor of tissue necrosis during acute inflammatory events. J. Exp. Med. 2008, 205, 2609–2621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Botos, I.; Wang, Y.; Leonard, J.N.; Shiloach, J.; Segal, D.M.; Davies, D.R. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science (N. Y.) 2008, 320, 379–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef]
- Adams, M.; Navabi, H.; Jasani, B.; Man, S.; Fiander, A.; Evans, A.S.; Donninger, C.; Mason, M. Dendritic cell (DC) based therapy for cervical cancer: Use of DC pulsed with tumour lysate and matured with a novel synthetic clinically non-toxic double stranded RNA analogue poly [I]:poly [C(12)U] (Ampligen R). Vaccine 2003, 21, 787–790. [Google Scholar] [CrossRef]
- Vanpouille-Box, C.; Hoffmann, J.A.; Galluzzi, L. Pharmacological modulation of nucleic acid sensors—therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2019, 18, 845–867. [Google Scholar] [CrossRef] [PubMed]
- Poltorak, A.; He, X.; Smirnova, I.; Liu, M.Y.; Van Huffel, C.; Du, X.; Birdwell, D.; Alejos, E.; Silva, M.; Galanos, C.; et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science (N. Y.) 1998, 282, 2085–2088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimazu, R.; Akashi, S.; Ogata, H.; Nagai, Y.; Fukudome, K.; Miyake, K.; Kimoto, M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 1999, 189, 1777–1782. [Google Scholar] [CrossRef]
- Biragyn, A.; Ruffini, P.A.; Leifer, C.A.; Klyushnenkova, E.; Shakhov, A.; Chertov, O.; Shirakawa, A.K.; Farber, J.M.; Segal, D.M.; Oppenheim, J.J.; et al. Toll-like receptor 4-dependent activation of dendritic cells by beta-defensin 2. Science (N. Y.) 2002, 298, 1025–1029. [Google Scholar] [CrossRef]
- Okamura, Y.; Watari, M.; Jerud, E.S.; Young, D.W.; Ishizaka, S.T.; Rose, J.; Chow, J.C.; Strauss, J.F., 3rd. The extra domain A of fibronectin activates Toll-like receptor 4. J. Boil. Chem. 2001, 276, 10229–10233. [Google Scholar] [CrossRef] [Green Version]
- Smiley, S.T.; King, J.A.; Hancock, W.W. Fibrinogen stimulates macrophage chemokine secretion through toll-like receptor 4. J. Immunol. (Baltim. Md. 1950) 2001, 167, 2887–2894. [Google Scholar] [CrossRef] [Green Version]
- Guillot, L.; Balloy, V.; McCormack, F.X.; Golenbock, D.T.; Chignard, M.; Si-Tahar, M. Cutting edge: The immunostimulatory activity of the lung surfactant protein-A involves Toll-like receptor 4. J. Immunol. (Baltim. Md. 1950) 2002, 168, 5989–5992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, Y.; Kuba, K.; Neely, G.G.; Yaghubian-Malhami, R.; Perkmann, T.; van Loo, G.; Ermolaeva, M.; Veldhuizen, R.; Leung, Y.H.; Wang, H.; et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 2008, 133, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.A.; Wheeler, D.S.; Lierl, K.M.; Hughes, V.S.; Wong, H.R.; Page, K. Hsp72 induces inflammation and regulates cytokine production in airway epithelium through a TLR4- and NF-kappaB-dependent mechanism. J. Immunol. (Baltim. Md. 1950) 2007, 179, 6318–6324. [Google Scholar] [CrossRef]
- Ohashi, K.; Burkart, V.; Flohe, S.; Kolb, H. Cutting edge: Heat shock protein 60 is a putative endogenous ligand of the toll-like receptor-4 complex. J. Immunol. (Baltim. Md. 1950) 2000, 164, 558–561. [Google Scholar] [CrossRef] [Green Version]
- Apetoh, L.; Ghiringhelli, F.; Tesniere, A.; Obeid, M.; Ortiz, C.; Criollo, A.; Mignot, G.; Maiuri, M.C.; Ullrich, E.; Saulnier, P.; et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 2007, 13, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Adanitsch, F.; Shi, J.; Shao, F.; Beyaert, R.; Heine, H.; Zamyatina, A. Synthetic glycan-based TLR4 agonists targeting caspase-4/11 for the development of adjuvants and immunotherapeutics. Chem. Sci. 2018, 9, 3957–3963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thoelen, S.; De Clercq, N.; Tornieporth, N. A prophylactic hepatitis B vaccine with a novel adjuvant system. Vaccine 2001, 19, 2400–2403. [Google Scholar] [CrossRef]
- Nascimento, E.; Fernandes, D.F.; Vieira, E.P.; Campos-Neto, A.; Ashman, J.A.; Alves, F.P.; Coler, R.N.; Bogatzki, L.Y.; Kahn, S.J.; Beckmann, A.M.; et al. A clinical trial to evaluate the safety and immunogenicity of the LEISH-F1+MPL-SE vaccine when used in combination with meglumine antimoniate for the treatment of cutaneous leishmaniasis. Vaccine 2010, 28, 6581–6587. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, F.; Yang, J.; Zhong, M.; Zhang, E.; Li, Y.; Zhou, D.; Cao, Y.; Li, W.; Yu, J.; et al. Over-activation of TLR5 signaling by high-dose flagellin induces liver injury in mice. Cell. Mol. Immunol. 2015, 12, 729–742. [Google Scholar] [CrossRef] [Green Version]
- Mizel, S.B.; Graff, A.H.; Sriranganathan, N.; Ervin, S.; Lees, C.J.; Lively, M.O.; Hantgan, R.R.; Thomas, M.J.; Wood, J.; Bell, B. Flagellin-F1-V fusion protein is an effective plague vaccine in mice and two species of nonhuman primates. Clin. Vaccine Immunol. 2009, 16, 21–28. [Google Scholar] [CrossRef] [Green Version]
- Carapau, D.; Mitchell, R.; Nacer, A.; Shaw, A.; Othoro, C.; Frevert, U.; Nardin, E. Protective humoral immunity elicited by a needle-free malaria vaccine comprised of a chimeric Plasmodium falciparum circumsporozoite protein and a Toll-like receptor 5 agonist, flagellin. Infect. Immun. 2013, 81, 4350–4362. [Google Scholar] [CrossRef] [Green Version]
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science (N. Y.) 2004, 303, 1529–1531. [Google Scholar] [CrossRef]
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science (N. Y.) 2004, 303, 1526–1529. [Google Scholar] [CrossRef] [Green Version]
- Gantier, M.P.; Tong, S.; Behlke, M.A.; Xu, D.; Phipps, S.; Foster, P.S.; Williams, B.R. TLR7 is involved in sequence-specific sensing of single-stranded RNAs in human macrophages. J. Immunol. (Baltim. Md. 1950) 2008, 180, 2117–2124. [Google Scholar] [CrossRef]
- Kelly, K.M.; Zhuang, H.; Nacionales, D.C.; Scumpia, P.O.; Lyons, R.; Akaogi, J.; Lee, P.; Williams, B.; Yamamoto, M.; Akira, S.; et al. “Endogenous adjuvant” activity of the RNA components of lupus autoantigens Sm/RNP and Ro 60. Arthritis Rheum 2006, 54, 1557–1567. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M. Innate sensing of self and non-self RNAs by Toll-like receptors. Trends Mol. Med. 2006, 12, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Van Hoeven, N.; Fox, C.B.; Granger, B.; Evers, T.; Joshi, S.W.; Nana, G.I.; Evans, S.C.; Lin, S.; Liang, H.; Liang, L.; et al. A Formulated TLR7/8 Agonist is a Flexible, Highly Potent and Effective Adjuvant for Pandemic Influenza Vaccines. Sci. Rep. 2017, 7, 46426. [Google Scholar] [CrossRef]
- Adams, S.; O’Neill, D.W.; Nonaka, D.; Hardin, E.; Chiriboga, L.; Siu, K.; Cruz, C.M.; Angiulli, A.; Angiulli, F.; Ritter, E.; et al. Immunization of malignant melanoma patients with full-length NY-ESO-1 protein using TLR7 agonist imiquimod as vaccine adjuvant. J. Immunol. (Baltim. Md. 1950) 2008, 181, 776–784. [Google Scholar] [CrossRef] [Green Version]
- Smorlesi, A.; Papalini, F.; Orlando, F.; Donnini, A.; Re, F.; Provinciali, M. Imiquimod and S-27609 as adjuvants of DNA vaccination in a transgenic murine model of HER2/neu-positive mammary carcinoma. Gene Ther. 2005, 12, 1324–1332. [Google Scholar] [CrossRef]
- Gordon, J.R.; Li, F.; Nayyar, A.; Xiang, J.; Zhang, X. CD8 alpha+, but not CD8 alpha-, dendritic cells tolerize Th2 responses via contact-dependent and -independent mechanisms, and reverse airway hyperresponsiveness, Th2, and eosinophil responses in a mouse model of asthma. J. Immunol. (Baltim. Md. 1950) 2005, 175, 1516–1522. [Google Scholar] [CrossRef] [Green Version]
- Ma, F.; Zhang, J.; Zhang, J.; Zhang, C. The TLR7 agonists imiquimod and gardiquimod improve DC-based immunotherapy for melanoma in mice. Cell. Mol. Immunol. 2010, 7, 381–388. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Kanuma, T.; Takahama, S.; Okamura, T.; Moriishi, E.; Ishii, K.J.; Terahara, K.; Yasutomi, Y. STING agonists activate latently infected cells and enhance SIV-specific responses ex vivo in naturally SIV controlled cynomolgus macaques. Sci. Rep. 2019, 9, 5917. [Google Scholar] [CrossRef]
- Demaria, O.; Pagni, P.P.; Traub, S.; de Gassart, A.; Branzk, N.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.D.; Flavell, R.A.; Alexopoulou, L. TLR8 deficiency leads to autoimmunity in mice. J. Clin. Investig. 2010, 120, 3651–3662. [Google Scholar] [CrossRef]
- Cervantes, J.L.; Weinerman, B.; Basole, C.; Salazar, J.C. TLR8: The forgotten relative revindicated. Cell. Mol. Immunol. 2012, 9, 434–438. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Cox, C.J.; Alvarez, K.M.; Cunningham, M.W. Cutting edge: Cardiac myosin activates innate immune responses through TLRs. J. Immunol. (Baltim. Md. 1950) 2009, 183, 27–31. [Google Scholar] [CrossRef] [Green Version]
- Takeshita, F.; Leifer, C.A.; Gursel, I.; Ishii, K.J.; Takeshita, S.; Gursel, M.; Klinman, D.M. Cutting edge: Role of Toll-like receptor 9 in CpG DNA-induced activation of human cells. J. Immunol. (Baltim. Md. 1950) 2001, 167, 3555–3558. [Google Scholar] [CrossRef] [Green Version]
- Murakami, Y.; Fukui, R.; Motoi, Y.; Shibata, T.; Saitoh, S.I.; Sato, R.; Miyake, K. The protective effect of the anti-Toll-like receptor 9 antibody against acute cytokine storm caused by immunostimulatory DNA. Sci. Rep. 2017, 7, 44042. [Google Scholar] [CrossRef]
- Leadbetter, E.A.; Rifkin, I.R.; Hohlbaum, A.M.; Beaudette, B.C.; Shlomchik, M.J.; Marshak-Rothstein, A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature 2002, 416, 603–607. [Google Scholar] [CrossRef]
- Viglianti, G.A.; Lau, C.M.; Hanley, T.M.; Miko, B.A.; Shlomchik, M.J.; Marshak-Rothstein, A. Activation of autoreactive B cells by CpG dsDNA. Immunity 2003, 19, 837–847. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Avalos, A.M.; Mao, S.Y.; Chen, B.; Senthil, K.; Wu, H.; Parroche, P.; Drabic, S.; Golenbock, D.; Sirois, C.; et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 2007, 8, 487–496. [Google Scholar] [CrossRef]
- Yeh, D.W.; Lai, C.Y.; Liu, Y.L.; Lu, C.H.; Tseng, P.H.; Yuh, C.H.; Yu, G.Y.; Liu, S.J.; Leng, C.H.; Chuang, T.H. CpG-oligodeoxynucleotides developed for grouper toll-like receptor (TLR) 21s effectively activate mouse and human TLR9s mediated immune responses. Sci. Rep. 2017, 7, 17297. [Google Scholar] [CrossRef] [Green Version]
- Cooper, C.L.; Davis, H.L.; Morris, M.L.; Efler, S.M.; Krieg, A.M.; Li, Y.; Laframboise, C.; Al Adhami, M.J.; Khaliq, Y.; Seguin, I.; et al. Safety and immunogenicity of CPG 7909 injection as an adjuvant to Fluarix influenza vaccine. Vaccine 2004, 22, 3136–3143. [Google Scholar] [CrossRef]
- Halperin, S.A.; Van Nest, G.; Smith, B.; Abtahi, S.; Whiley, H.; Eiden, J.J. A phase I study of the safety and immunogenicity of recombinant hepatitis B surface antigen co-administered with an immunostimulatory phosphorothioate oligonucleotide adjuvant. Vaccine 2003, 21, 2461–2467. [Google Scholar] [CrossRef]
- Oosting, M.; Cheng, S.C.; Bolscher, J.M.; Vestering-Stenger, R.; Plantinga, T.S.; Verschueren, I.C.; Arts, P.; Garritsen, A.; van Eenennaam, H.; Sturm, P.; et al. Human TLR10 is an anti-inflammatory pattern-recognition receptor. Proc. Natl. Acad. Sci. USA 2014, 111, E4478–E4484. [Google Scholar] [CrossRef] [Green Version]
- Henrick, B.M.; Yao, X.D.; Zahoor, M.A.; Abimiku, A.; Osawe, S.; Rosenthal, K.L. TLR10 Senses HIV-1 Proteins and Significantly Enhances HIV-1 Infection. Front. Immunol. 2019, 10, 482. [Google Scholar] [CrossRef]
- Raetz, M.; Kibardin, A.; Sturge, C.R.; Pifer, R.; Li, H.; Burstein, E.; Ozato, K.; Larin, S.; Yarovinsky, F. Cooperation of TLR12 and TLR11 in the IRF8-dependent IL-12 response to Toxoplasma gondii profilin. J. Immunol. (Baltim. Md. 1950) 2013, 191, 4818–4827. [Google Scholar] [CrossRef] [Green Version]
- Koblansky, A.A.; Jankovic, D.; Oh, H.; Hieny, S.; Sungnak, W.; Mathur, R.; Hayden, M.S.; Akira, S.; Sher, A.; Ghosh, S. Recognition of profilin by Toll-like receptor 12 is critical for host resistance to Toxoplasma gondii. Immunity 2013, 38, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Wang, C.; Cheng, P.; Zhang, X.; Li, X.; Hu, Y.; Xu, F.; Hong, F.; Dong, G.; Xiong, H. CD180 Ligation Inhibits TLR7- and TLR9-Mediated Activation of Macrophages and Dendritic Cells Through the Lyn-SHP-1/2 Axis in Murine Lupus. Front. Immunol. 2018, 9, 2643. [Google Scholar] [CrossRef]
- Ramanjulu, J.M.; Pesiridis, G.S.; Yang, J.; Concha, N.; Singhaus, R.; Zhang, S.Y.; Tran, J.L.; Moore, P.; Lehmann, S.; Eberl, H.C.; et al. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature 2018, 564, 439–443. [Google Scholar] [CrossRef]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Naslund, T.I.; Liljestrom, P.; Weber, F.; Reis e Sousa, C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science (N. Y.) 2006, 314, 997–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, K.; Du, J.; Peng, Y.; Li, P.; Wang, S.; Wang, Y.; Hou, J.; Kang, J.; Zheng, W.; Hua, S.; et al. LINE1 contributes to autoimmunity through both RIG-I- and MDA5-mediated RNA sensing pathways. J. Autoimmun. 2018, 90, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ye, X.; Dunker, W.; Song, Y.; Karijolich, J. RIG-I like receptor sensing of host RNAs facilitates the cell-intrinsic immune response to KSHV infection. Nat. Commun. 2018, 9, 4841. [Google Scholar] [CrossRef] [PubMed]
- Kasumba, D.M.; Grandvaux, N. Therapeutic Targeting of RIG-I and MDA5 Might Not Lead to the Same Rome. Trends Pharm. Sci. 2019, 40, 116–127. [Google Scholar] [CrossRef]
- Linehan, M.M.; Dickey, T.H.; Molinari, E.S.; Fitzgerald, M.E.; Potapova, O.; Iwasaki, A.; Pyle, A.M. A minimal RNA ligand for potent RIG-I activation in living mice. Sci. Adv. 2018, 4, e1701854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias Junior, A.G.; Sampaio, N.G.; Rehwinkel, J. A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. Trends Microbiol. 2019, 27, 75–85. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.; Mu, X.; Yang, F.; Greenwald, E.; Park, J.W.; Jacob, E.; Zhang, C.Z.; Hur, S. Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation. Cell 2018, 172, 797–810.e713. [Google Scholar] [CrossRef] [Green Version]
- Dhir, A.; Dhir, S.; Borowski, L.S.; Jimenez, L.; Teitell, M.; Rotig, A.; Crow, Y.J.; Rice, G.I.; Duffy, D.; Tamby, C.; et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 2018, 560, 238–242. [Google Scholar] [CrossRef]
- Roulois, D.; Loo Yau, H.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Jones, P.A.; Pugh, T.J.; et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef] [Green Version]
- Bevan, M.J. Helping the CD8(+) T-cell response. Nat. Rev. Immunol. 2004, 4, 595–602. [Google Scholar] [CrossRef]
- Borst, J.; Ahrends, T.; Babala, N.; Melief, C.J.M.; Kastenmuller, W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018, 18, 635–647. [Google Scholar] [CrossRef]
- Janssen, E.M.; Lemmens, E.E.; Wolfe, T.; Christen, U.; von Herrath, M.G.; Schoenberger, S.P. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature 2003, 421, 852–856. [Google Scholar] [CrossRef]
- Laidlaw, B.J.; Craft, J.E.; Kaech, S.M. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat. Rev. Immunol. 2016, 16, 102–111. [Google Scholar] [CrossRef]
- Shedlock, D.J.; Shen, H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science (N. Y.) 2003, 300, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Hunder, N.N.; Wallen, H.; Cao, J.; Hendricks, D.W.; Reilly, J.Z.; Rodmyre, R.; Jungbluth, A.; Gnjatic, S.; Thompson, J.A.; Yee, C. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N. Engl. J. Med. 2008, 358, 2698–2703. [Google Scholar] [CrossRef] [Green Version]
- Tran, E.; Turcotte, S.; Gros, A.; Robbins, P.F.; Lu, Y.C.; Dudley, M.E.; Wunderlich, J.R.; Somerville, R.P.; Hogan, K.; Hinrichs, C.S.; et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science (N. Y.) 2014, 344, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Billings, P.; Burakoff, S.; Dorf, M.E.; Benacerraf, B. Cytotoxic T lymphocytes specific for I region determinants do not require interactions with H-2K or D gene products. J. Exp. Med. 1977, 145, 1387–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, A.; Saito, T. CD4 CTL, a Cytotoxic Subset of CD4(+) T Cells, Their Differentiation and Function. Front. Immunol. 2017, 8, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, H.; Gotze, D.; Ptschelinzew, L.; Rollinghoff, M. Induction of cytotoxic T lymphocytes against I-region-coded determinants: In vitro evidence for a third histocompatibility locus in the mouse. J. Exp. Med. 1975, 142, 1477–1487. [Google Scholar] [CrossRef]
- Quezada, S.A.; Simpson, T.R.; Peggs, K.S.; Merghoub, T.; Vider, J.; Fan, X.; Blasberg, R.; Yagita, H.; Muranski, P.; Antony, P.A.; et al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J. Exp. Med. 2010, 207, 637–650. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Akpinarli, A.; Maris, C.; Hipkiss, E.L.; Lane, M.; Kwon, E.K.; Muranski, P.; Restifo, N.P.; Antony, P.A. Naive tumor-specific CD4(+) T cells differentiated in vivo eradicate established melanoma. J. Exp. Med. 2010, 207, 651–667. [Google Scholar] [CrossRef] [Green Version]
- Eickhoff, S.; Brewitz, A.; Gerner, M.Y.; Klauschen, F.; Komander, K.; Hemmi, H.; Garbi, N.; Kaisho, T.; Germain, R.N.; Kastenmuller, W. Robust Anti-viral Immunity Requires Multiple Distinct T Cell-Dendritic Cell Interactions. Cell 2015, 162, 1322–1337. [Google Scholar] [CrossRef] [Green Version]
- Hor, J.L.; Whitney, P.G.; Zaid, A.; Brooks, A.G.; Heath, W.R.; Mueller, S.N. Spatiotemporally Distinct Interactions with Dendritic Cell Subsets Facilitates CD4+ and CD8+ T Cell Activation to Localized Viral Infection. Immunity 2015, 43, 554–565. [Google Scholar] [CrossRef] [Green Version]
- Allan, R.S.; Waithman, J.; Bedoui, S.; Jones, C.M.; Villadangos, J.A.; Zhan, Y.; Lew, A.M.; Shortman, K.; Heath, W.R.; Carbone, F.R. Migratory dendritic cells transfer antigen to a lymph node-resident dendritic cell population for efficient CTL priming. Immunity 2006, 25, 153–162. [Google Scholar] [CrossRef] [Green Version]
- Schoenberger, S.P.; Toes, R.E.; van der Voort, E.I.; Offringa, R.; Melief, C.J. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature 1998, 393, 480–483. [Google Scholar] [CrossRef]
- Sun, J.C.; Bevan, M.J. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science (N. Y.) 2003, 300, 339–342. [Google Scholar] [CrossRef] [Green Version]
- Ahrends, T.; Spanjaard, A.; Pilzecker, B.; Babala, N.; Bovens, A.; Xiao, Y.; Jacobs, H.; Borst, J. CD4(+) T Cell Help Confers a Cytotoxic T Cell Effector Program Including Coinhibitory Receptor Downregulation and Increased Tissue Invasiveness. Immunity 2017, 47, 848–861.e845. [Google Scholar] [CrossRef] [Green Version]
- Bennett, S.R.; Carbone, F.R.; Karamalis, F.; Flavell, R.A.; Miller, J.F.; Heath, W.R. Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature 1998, 393, 478–480. [Google Scholar] [CrossRef]
- Calabro, S.; Liu, D.; Gallman, A.; Nascimento, M.S.; Yu, Z.; Zhang, T.T.; Chen, P.; Zhang, B.; Xu, L.; Gowthaman, U.; et al. Differential Intrasplenic Migration of Dendritic Cell Subsets Tailors Adaptive Immunity. Cell Rep. 2016, 16, 2472–2485. [Google Scholar] [CrossRef] [Green Version]
- Castellino, F.; Huang, A.Y.; Altan-Bonnet, G.; Stoll, S.; Scheinecker, C.; Germain, R.N. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature 2006, 440, 890–895. [Google Scholar] [CrossRef]
- Ridge, J.P.; Di Rosa, F.; Matzinger, P. A conditioned dendritic cell can be a temporal bridge between a CD4+ T-helper and a T-killer cell. Nature 1998, 393, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Baptista, A.P.; Gola, A.; Huang, Y.; Milanez-Almeida, P.; Torabi-Parizi, P.; Urban, J.F., Jr.; Shapiro, V.S.; Gerner, M.Y.; Germain, R.N. The Chemoattractant Receptor Ebi2 Drives Intranodal Naive CD4(+) T Cell Peripheralization to Promote Effective Adaptive Immunity. Immunity 2019, 50, 1188–1201.e1186. [Google Scholar] [CrossRef] [PubMed]
- Gerner, M.Y.; Kastenmuller, W.; Ifrim, I.; Kabat, J.; Germain, R.N. Histo-cytometry: A method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 2012, 37, 364–376. [Google Scholar] [CrossRef] [Green Version]
- Hoyer, S.; Prommersberger, S.; Pfeiffer, I.A.; Schuler-Thurner, B.; Schuler, G.; Dorrie, J.; Schaft, N. Concurrent interaction of DCs with CD4(+) and CD8(+) T cells improves secondary CTL expansion: It takes three to tango. Eur. J. Immunol. 2014, 44, 3543–3559. [Google Scholar] [CrossRef]
- Sokke Umeshappa, C.; Hebbandi Nanjundappa, R.; Xie, Y.; Freywald, A.; Deng, Y.; Ma, H.; Xiang, J. CD154 and IL-2 signaling of CD4+ T cells play a critical role in multiple phases of CD8+ CTL responses following adenovirus vaccination. PLoS ONE 2012, 7, e47004. [Google Scholar] [CrossRef] [Green Version]
- Wilson, E.B.; Livingstone, A.M. Cutting edge: CD4+ T cell-derived IL-2 is essential for help-dependent primary CD8+ T cell responses. J. Immunol. (Baltim. Md. 1950) 2008, 181, 7445–7448. [Google Scholar] [CrossRef] [Green Version]
- Provine, N.M.; Larocca, R.A.; Aid, M.; Penaloza-MacMaster, P.; Badamchi-Zadeh, A.; Borducchi, E.N.; Yates, K.B.; Abbink, P.; Kirilova, M.; Ng’ang’a, D.; et al. Immediate Dysfunction of Vaccine-Elicited CD8+ T Cells Primed in the Absence of CD4+ T Cells. J. Immunol. (Baltim. Md. 1950) 2016, 197, 1809–1822. [Google Scholar] [CrossRef] [Green Version]
- Nakanishi, Y.; Lu, B.; Gerard, C.; Iwasaki, A. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature 2009, 462, 510–513. [Google Scholar] [CrossRef] [Green Version]
- Spitzer, M.H.; Carmi, Y.; Reticker-Flynn, N.E.; Kwek, S.S.; Madhireddy, D.; Martins, M.M.; Gherardini, P.F.; Prestwood, T.R.; Chabon, J.; Bendall, S.C.; et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 2017, 168, 487–502. [Google Scholar] [CrossRef]
- Togashi, Y.; Shitara, K.; Nishikawa, H. Regulatory T cells in cancer immunosuppression—implications for anticancer therapy. Nat. Rev. Clin. Oncol. 2019, 16, 356–371. [Google Scholar] [CrossRef]
- Plitas, G.; Rudensky, A.Y. Regulatory T Cells in Cancer. Annu. Rev. Cancer Biol. 2020, 4, 459–477. [Google Scholar] [CrossRef] [Green Version]
- Kalia, V.; Penny, L.A.; Yuzefpolskiy, Y.; Baumann, F.M.; Sarkar, S. Quiescence of Memory CD8(+) T Cells Is Mediated by Regulatory T Cells through Inhibitory Receptor CTLA-4. Immunity 2015, 42, 1116–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pace, L.; Tempez, A.; Arnold-Schrauf, C.; Lemaitre, F.; Bousso, P.; Fetler, L.; Sparwasser, T.; Amigorena, S. Regulatory T cells increase the avidity of primary CD8+ T cell responses and promote memory. Science (N. Y.) 2012, 338, 532–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtsinger, J.M.; Lins, D.C.; Mescher, M.F. Signal 3 determines tolerance versus full activation of naive CD8 T cells: Dissociating proliferation and development of effector function. J. Exp. Med. 2003, 197, 1141–1151. [Google Scholar] [CrossRef]
- Curtsinger, J.M.; Schmidt, C.S.; Mondino, A.; Lins, D.C.; Kedl, R.M.; Jenkins, M.K.; Mescher, M.F. Inflammatory cytokines provide a third signal for activation of naive CD4+ and CD8+ T cells. J. Immunol. (Baltim. Md. 1950) 1999, 162, 3256–3262. [Google Scholar]
- Curtsinger, J.M.; Valenzuela, J.O.; Agarwal, P.; Lins, D.; Mescher, M.F. Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J. Immunol. (Baltim. Md. 1950) 2005, 174, 4465–4469. [Google Scholar] [CrossRef] [Green Version]
- Ehrich, E.W.; Devaux, B.; Rock, E.P.; Jorgensen, J.L.; Davis, M.M.; Chien, Y.H. T cell receptor interaction with peptide/major histocompatibility complex (MHC) and superantigen/MHC ligands is dominated by antigen. J. Exp. Med. 1993, 178, 713–722. [Google Scholar] [CrossRef]
- Wu, L.C.; Tuot, D.S.; Lyons, D.S.; Garcia, K.C.; Davis, M.M. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature 2002, 418, 552–556. [Google Scholar] [CrossRef]
- Henrickson, S.E.; Mempel, T.R.; Mazo, I.B.; Liu, B.; Artyomov, M.N.; Zheng, H.; Peixoto, A.; Flynn, M.P.; Senman, B.; Junt, T.; et al. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat. Immunol. 2008, 9, 282–291. [Google Scholar] [CrossRef] [Green Version]
- Henrickson, S.E.; Perro, M.; Loughhead, S.M.; Senman, B.; Stutte, S.; Quigley, M.; Alexe, G.; Iannacone, M.; Flynn, M.P.; Omid, S.; et al. Antigen availability determines CD8(+) T cell-dendritic cell interaction kinetics and memory fate decisions. Immunity 2013, 39, 496–507. [Google Scholar] [CrossRef] [Green Version]
- Manz, B.N.; Jackson, B.L.; Petit, R.S.; Dustin, M.L.; Groves, J. T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proc. Natl. Acad. Sci. USA 2011, 108, 9089–9094. [Google Scholar] [CrossRef] [Green Version]
- Bedoui, S.; Whitney, P.G.; Waithman, J.; Eidsmo, L.; Wakim, L.; Caminschi, I.; Allan, R.S.; Wojtasiak, M.; Shortman, K.; Carbone, F.R.; et al. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat. Immunol. 2009, 10, 488–495. [Google Scholar] [CrossRef]
- Jongbloed, S.L.; Kassianos, A.J.; McDonald, K.J.; Clark, G.J.; Ju, X.; Angel, C.E.; Chen, C.J.; Dunbar, P.R.; Wadley, R.B.; Jeet, V.; et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 2010, 207, 1247–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appleman, L.J.; Berezovskaya, A.; Grass, I.; Boussiotis, V.A. CD28 costimulation mediates T cell expansion via IL-2-independent and IL-2-dependent regulation of cell cycle progression. J. Immunol. (Baltim. Md. 1950) 2000, 164, 144–151. [Google Scholar] [CrossRef] [Green Version]
- Krummel, M.F.; Allison, J.P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 1995, 182, 459–465. [Google Scholar] [CrossRef] [Green Version]
- Marengere, L.E.; Waterhouse, P.; Duncan, G.S.; Mittrucker, H.W.; Feng, G.S.; Mak, T.W. Regulation of T cell receptor signaling by tyrosine phosphatase SYP association with CTLA-4. Science (N. Y.) 1996, 272, 1170–1173. [Google Scholar] [CrossRef]
- van der Merwe, P.A.; Bodian, D.L.; Daenke, S.; Linsley, P.; Davis, S.J. CD80 (B7-1) binds both CD28 and CTLA-4 with a low affinity and very fast kinetics. J. Exp. Med. 1997, 185, 393–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, P.; Penninger, J.M.; Timms, E.; Wakeham, A.; Shahinian, A.; Lee, K.P.; Thompson, C.B.; Griesser, H.; Mak, T.W. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science (N. Y.) 1995, 270, 985–988. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.V.; Brodie, D.W.; Gilbert, R.J.; Iaboni, A.; Manso-Sancho, R.; Walse, B.; Stuart, D.I.; van der Merwe, P.A.; Davis, S.J. The interaction properties of costimulatory molecules revisited. Immunity 2002, 17, 201–210. [Google Scholar] [CrossRef] [Green Version]
- Rowshanravan, B.; Halliday, N.; Sansom, D.M. CTLA-4: A moving target in immunotherapy. Blood 2018, 131, 58–67. [Google Scholar] [CrossRef]
- Schwartz, J.C.; Zhang, X.; Fedorov, A.A.; Nathenson, S.G.; Almo, S.C. Structural basis for co-stimulation by the human CTLA-4/B7-2 complex. Nature 2001, 410, 604–608. [Google Scholar] [CrossRef]
- Linsley, P.S.; Bradshaw, J.; Greene, J.; Peach, R.; Bennett, K.L.; Mittler, R.S. Intracellular trafficking of CTLA-4 and focal localization towards sites of TCR engagement. Immunity 1996, 4, 535–543. [Google Scholar] [CrossRef] [Green Version]
- Shiratori, T.; Miyatake, S.; Ohno, H.; Nakaseko, C.; Isono, K.; Bonifacino, J.S.; Saito, T. Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity 1997, 6, 583–589. [Google Scholar] [CrossRef] [Green Version]
- Walunas, T.L.; Lenschow, D.J.; Bakker, C.Y.; Linsley, P.S.; Freeman, G.J.; Green, J.M.; Thompson, C.B.; Bluestone, J.A. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1994, 1, 405–413. [Google Scholar] [CrossRef]
- Qureshi, O.S.; Zheng, Y.; Nakamura, K.; Attridge, K.; Manzotti, C.; Schmidt, E.M.; Baker, J.; Jeffery, L.E.; Kaur, S.; Briggs, Z.; et al. Trans-endocytosis of CD80 and CD86: A molecular basis for the cell-extrinsic function of CTLA-4. Science (N. Y.) 2011, 332, 600–603. [Google Scholar] [CrossRef] [Green Version]
- Leach, D.R.; Krummel, M.F.; Allison, J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science (N. Y.) 1996, 271, 1734–1736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingram, J.R.; Blomberg, O.S.; Rashidian, M.; Ali, L.; Garforth, S.; Fedorov, E.; Fedorov, A.A.; Bonanno, J.B.; Le Gall, C.; Crowley, S.; et al. Anti-CTLA-4 therapy requires an Fc domain for efficacy. Proc. Natl. Acad. Sci. USA 2018, 115, 3912–3917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramagopal, U.A.; Liu, W.; Garrett-Thomson, S.C.; Bonanno, J.B.; Yan, Q.; Srinivasan, M.; Wong, S.C.; Bell, A.; Mankikar, S.; Rangan, V.S.; et al. Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab. Proc. Natl. Acad. Sci. USA 2017, 114, E4223–E4232. [Google Scholar] [CrossRef] [Green Version]
- Read, S.; Greenwald, R.; Izcue, A.; Robinson, N.; Mandelbrot, D.; Francisco, L.; Sharpe, A.H.; Powrie, F. Blockade of CTLA-4 on CD4+CD25+ regulatory T cells abrogates their function in vivo. J. Immunol. (Baltim. Md. 1950) 2006, 177, 4376–4383. [Google Scholar] [CrossRef] [Green Version]
- Simpson, T.R.; Li, F.; Montalvo-Ortiz, W.; Sepulveda, M.A.; Bergerhoff, K.; Arce, F.; Roddie, C.; Henry, J.Y.; Yagita, H.; Wolchok, J.D.; et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J. Exp. Med. 2013, 210, 1695–1710. [Google Scholar] [CrossRef] [PubMed]
- Wing, K.; Onishi, Y.; Prieto-Martin, P.; Yamaguchi, T.; Miyara, M.; Fehervari, Z.; Nomura, T.; Sakaguchi, S. CTLA-4 control over Foxp3+ regulatory T cell function. Science (N. Y.) 2008, 322, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Boutros, C.; Tarhini, A.; Routier, E.; Lambotte, O.; Ladurie, F.L.; Carbonnel, F.; Izzeddine, H.; Marabelle, A.; Champiat, S.; Berdelou, A.; et al. Safety profiles of anti-CTLA-4 and anti-PD-1 antibodies alone and in combination. Nat. Rev. Clin. Oncol. 2016, 13, 473–486. [Google Scholar] [CrossRef]
- Verhagen, J.; Genolet, R.; Britton, G.J.; Stevenson, B.J.; Sabatos-Peyton, C.A.; Dyson, J.; Luescher, I.F.; Wraith, D.C. CTLA-4 controls the thymic development of both conventional and regulatory T cells through modulation of the TCR repertoire. Proc. Natl. Acad. Sci. USA 2013, 110, E221–E230. [Google Scholar] [CrossRef] [Green Version]
- Vetizou, M.; Pitt, J.M.; Daillere, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (N. Y.) 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [Green Version]
- Zenke, S.; Palm, M.M.; Braun, J.; Gavrilov, A.; Meiser, P.; Bottcher, J.P.; Beyersdorf, N.; Ehl, S.; Gerard, A.; Lammermann, T.; et al. Quorum Regulation via Nested Antagonistic Feedback Circuits Mediated by the Receptors CD28 and CTLA-4 Confers Robustness to T Cell Population Dynamics. Immunity 2020, 52, 313–327.e317. [Google Scholar] [CrossRef]
- Gu, P.; Gao, J.F.; D’Souza, C.A.; Kowalczyk, A.; Chou, K.Y.; Zhang, L. Trogocytosis of CD80 and CD86 by induced regulatory T cells. Cell Mol. Immunol. 2012, 9, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Tatari-Calderone, Z.; Semnani, R.T.; Nutman, T.B.; Schlom, J.; Sabzevari, H. Acquisition of CD80 by human T cells at early stages of activation: Functional involvement of CD80 acquisition in T cell to T cell interaction. J. Immunol. (Baltim. Md. 1950) 2002, 169, 6162–6169. [Google Scholar] [CrossRef]
- Aicher, A.; Hayden-Ledbetter, M.; Brady, W.A.; Pezzutto, A.; Richter, G.; Magaletti, D.; Buckwalter, S.; Ledbetter, J.A.; Clark, E.A. Characterization of human inducible costimulator ligand expression and function. J. Immunol. (Baltim. Md. 1950) 2000, 164, 4689–4696. [Google Scholar] [CrossRef] [Green Version]
- Aspord, C.; Leccia, M.T.; Charles, J.; Plumas, J. Plasmacytoid dendritic cells support melanoma progression by promoting Th2 and regulatory immunity through OX40L and ICOSL. Cancer Immunol. Res. 2013, 1, 402–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gariepy, J.; Prodeus, A.; Sparkes, A.; Fischer, N.; Saha, S. A powerful ICOS agonist that enhances anti-tumor immune responses restored by immune checkpoint inhibitors. J. Immunol. 2019, 202, 71–75. [Google Scholar]
- Hutloff, A.; Dittrich, A.M.; Beier, K.C.; Eljaschewitsch, B.; Kraft, R.; Anagnostopoulos, I.; Kroczek, R.A. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature 1999, 397, 263–266. [Google Scholar] [CrossRef]
- Mages, H.W.; Hutloff, A.; Heuck, C.; Buchner, K.; Himmelbauer, H.; Oliveri, F.; Kroczek, R.A. Molecular cloning and characterization of murine ICOS and identification of B7h as ICOS ligand. Eur. J. Immunol. 2000, 30, 1040–1047. [Google Scholar] [CrossRef]
- Wikenheiser, D.J.; Stumhofer, J.S. ICOS Co-Stimulation: Friend or Foe? Front. Immunol. 2016, 7, 304. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Takagi, Y.; Kotani, M.; Hara, Y.; Inamine, A.; Hayashi, K.; Ogawa, S.; Takeda, K.; Tanabe, K.; Abe, R. Down-regulation of ICOS ligand by interaction with ICOS functions as a regulatory mechanism for immune responses. J. Immunol. (Baltim. Md. 1950) 2008, 180, 5222–5234. [Google Scholar] [CrossRef] [Green Version]
- Hedl, M.; Lahiri, A.; Ning, K.; Cho, J.H.; Abraham, C. Pattern recognition receptor signaling in human dendritic cells is enhanced by ICOS ligand and modulated by the Crohn’s disease ICOSLG risk allele. Immunity 2014, 40, 734–746. [Google Scholar] [CrossRef] [Green Version]
- Busse, M.; Krech, M.; Meyer-Bahlburg, A.; Hennig, C.; Hansen, G. ICOS mediates the generation and function of CD4+CD25+Foxp3+ regulatory T cells conveying respiratory tolerance. J. Immunol. (Baltim. Md. 1950) 2012, 189, 1975–1982. [Google Scholar] [CrossRef] [Green Version]
- Redpath, S.A.; van der Werf, N.; Cervera, A.M.; MacDonald, A.S.; Gray, D.; Maizels, R.M.; Taylor, M.D. ICOS controls Foxp3(+) regulatory T-cell expansion, maintenance and IL-10 production during helminth infection. Eur. J. Immunol. 2013, 43, 705–715. [Google Scholar] [CrossRef] [Green Version]
- Faget, J.; Bendriss-Vermare, N.; Gobert, M.; Durand, I.; Olive, D.; Biota, C.; Bachelot, T.; Treilleux, I.; Goddard-Leon, S.; Lavergne, E.; et al. ICOS-ligand expression on plasmacytoid dendritic cells supports breast cancer progression by promoting the accumulation of immunosuppressive CD4+ T cells. Cancer Res. 2012, 72, 6130–6141. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Quezada, S.A.; Sepulveda, M.A.; Sharma, P.; Allison, J.P. Engagement of the ICOS pathway markedly enhances efficacy of CTLA-4 blockade in cancer immunotherapy. J. Exp. Med. 2014, 211, 715–725. [Google Scholar] [CrossRef] [Green Version]
- Carthon, B.C.; Wolchok, J.D.; Yuan, J.; Kamat, A.; Ng Tang, D.S.; Sun, J.; Ku, G.; Troncoso, P.; Logothetis, C.J.; Allison, J.P.; et al. Preoperative CTLA-4 blockade: Tolerability and immune monitoring in the setting of a presurgical clinical trial. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 2861–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Liakou, C.I.; Kamat, A.; Pettaway, C.; Ward, J.F.; Tang, D.N.; Sun, J.; Jungbluth, A.A.; Troncoso, P.; Logothetis, C.; et al. Anti-CTLA-4 therapy results in higher CD4+ICOShi T cell frequency and IFN-gamma levels in both nonmalignant and malignant prostate tissues. Proc. Natl. Acad. Sci. USA 2009, 106, 2729–2734. [Google Scholar] [CrossRef] [Green Version]
- Liakou, C.I.; Kamat, A.; Tang, D.N.; Chen, H.; Sun, J.; Troncoso, P.; Logothetis, C.; Sharma, P. CTLA-4 blockade increases IFNgamma-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc. Natl. Acad. Sci. USA 2008, 105, 14987–14992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng Tang, D.; Shen, Y.; Sun, J.; Wen, S.; Wolchok, J.D.; Yuan, J.; Allison, J.P.; Sharma, P. Increased frequency of ICOS+ CD4 T cells as a pharmacodynamic biomarker for anti-CTLA-4 therapy. Cancer Immunol. Res. 2013, 1, 229–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Fu, T.; Suh, W.K.; Tsavachidou, D.; Wen, S.; Gao, J.; Ng Tang, D.; He, Q.; Sun, J.; Sharma, P. CD4 T cells require ICOS-mediated PI3K signaling to increase T-Bet expression in the setting of anti-CTLA-4 therapy. Cancer Immunol. Res. 2014, 2, 167–176. [Google Scholar] [CrossRef] [Green Version]
- Saoulli, K.; Lee, S.Y.; Cannons, J.L.; Yeh, W.C.; Santana, A.; Goldstein, M.D.; Bangia, N.; DeBenedette, M.A.; Mak, T.W.; Choi, Y.; et al. CD28-independent, TRAF2-dependent costimulation of resting T cells by 4-1BB ligand. J. Exp. Med. 1998, 187, 1849–1862. [Google Scholar] [CrossRef]
- Vezys, V.; Penaloza-MacMaster, P.; Barber, D.L.; Ha, S.J.; Konieczny, B.; Freeman, G.J.; Mittler, R.S.; Ahmed, R. 4-1BB signaling synergizes with programmed death ligand 1 blockade to augment CD8 T cell responses during chronic viral infection. J. Immunol. (Baltim. Md. 1950) 2011, 187, 1634–1642. [Google Scholar] [CrossRef]
- Placke, T.; Kopp, H.G.; Salih, H.R. Glucocorticoid-induced TNFR-related (GITR) protein and its ligand in antitumor immunity: Functional role and therapeutic modulation. Clin. Dev. Immunol. 2010, 2010, 239083. [Google Scholar] [CrossRef]
- Gurney, A.L.; Marsters, S.A.; Huang, R.M.; Pitti, R.M.; Mark, D.T.; Baldwin, D.T.; Gray, A.M.; Dowd, A.D.; Brush, A.D.; Heldens, A.D.; et al. Identification of a new member of the tumor necrosis factor family and its receptor, a human ortholog of mouse GITR. Curr. Boil. CB 1999, 9, 215–218. [Google Scholar] [CrossRef] [Green Version]
- Kamimura, Y.; Iwai, H.; Piao, J.; Hashiguchi, M.; Azuma, M. The glucocorticoid-induced TNF receptor-related protein (GITR)-GITR ligand pathway acts as a mediator of cutaneous dendritic cell migration and promotes T cell-mediated acquired immunity. J. Immunol. (Baltim. Md. 1950) 2009, 182, 2708–2716. [Google Scholar] [CrossRef] [Green Version]
- Kwon, B.; Yu, K.Y.; Ni, J.; Yu, G.L.; Jang, I.K.; Kim, Y.J.; Xing, L.; Liu, D.; Wang, S.X.; Kwon, B.S. Identification of a novel activation-inducible protein of the tumor necrosis factor receptor superfamily and its ligand. J. Boil. Chem. 1999, 274, 6056–6061. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.; Lee, S.; Lee, W.H.; Lee, H.J.; Suk, K. Stimulation of glucocorticoid-induced tumor necrosis factor receptor family-related protein ligand (GITRL) induces inflammatory activation of microglia in culture. J. Neurosci. Res. 2010, 88, 2188–2196. [Google Scholar] [CrossRef]
- Kim, J.D.; Choi, B.K.; Bae, J.S.; Lee, U.H.; Han, I.S.; Lee, H.W.; Youn, B.S.; Vinay, D.S.; Kwon, B.S. Cloning and characterization of GITR ligand. Genes Immun. 2003, 4, 564–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McHugh, R.S.; Whitters, M.J.; Piccirillo, C.A.; Young, D.A.; Shevach, E.M.; Collins, M.; Byrne, M.C. CD4(+)CD25(+) immunoregulatory T cells: Gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity 2002, 16, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Suvas, S.; Kim, B.; Sarangi, P.P.; Tone, M.; Waldmann, H.; Rouse, B.T. In vivo kinetics of GITR and GITR ligand expression and their functional significance in regulating viral immunopathology. J. Virol. 2005, 79, 11935–11942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tone, M.; Tone, Y.; Adams, E.; Yates, S.F.; Frewin, M.R.; Cobbold, S.P.; Waldmann, H. Mouse glucocorticoid-induced tumor necrosis factor receptor ligand is costimulatory for T cells. Proc. Natl. Acad. Sci. USA 2003, 100, 15059–15064. [Google Scholar] [CrossRef] [Green Version]
- Ji, H.B.; Liao, G.; Faubion, W.A.; Abadia-Molina, A.C.; Cozzo, C.; Laroux, F.S.; Caton, A.; Terhorst, C. Cutting edge: The natural ligand for glucocorticoid-induced TNF receptor-related protein abrogates regulatory T cell suppression. J. Immunol. (Baltim. Md. 1950) 2004, 172, 5823–5827. [Google Scholar] [CrossRef] [Green Version]
- Stephens, G.L.; McHugh, R.S.; Whitters, M.J.; Young, D.A.; Luxenberg, D.; Carreno, B.M.; Collins, M.; Shevach, E.M. Engagement of glucocorticoid-induced TNFR family-related receptor on effector T cells by its ligand mediates resistance to suppression by CD4+CD25+ T cells. J. Immunol. (Baltim. Md. 1950) 2004, 173, 5008–5020. [Google Scholar] [CrossRef] [Green Version]
- Vecchiarelli, A.; Pericolini, E.; Gabrielli, E.; Agostini, M.; Bistoni, F.; Nocentini, G.; Cenci, E.; Riccardi, C. The GITRL-GITR system alters TLR-4 expression on DC during fungal infection. Cell. Immunol. 2009, 257, 13–22. [Google Scholar] [CrossRef]
- Tuyaerts, S.; Van Meirvenne, S.; Bonehill, A.; Heirman, C.; Corthals, J.; Waldmann, H.; Breckpot, K.; Thielemans, K.; Aerts, J.L. Expression of human GITRL on myeloid dendritic cells enhances their immunostimulatory function but does not abrogate the suppressive effect of CD4+CD25+ regulatory T cells. J. Leukoc. Boil. 2007, 82, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.S.; Hsu, J.V.; Morrison, S.L. Localized expression of GITR-L in the tumor microenvironment promotes CD8+ T cell dependent anti-tumor immunity. Cancer Immunol. Immunother. 2009, 58, 1057–1069. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.D.; Diab, A.; Perales, M.A.; Wolchok, J.D.; Rizzuto, G.; Merghoub, T.; Huggins, D.; Liu, C.; Turk, M.J.; Restifo, N.P.; et al. Agonist anti-GITR antibody enhances vaccine-induced CD8(+) T-cell responses and tumor immunity. Cancer Res. 2006, 66, 4904–4912. [Google Scholar] [CrossRef] [Green Version]
- Durham, N.M.; Holoweckyj, N.; MacGill, R.S.; McGlinchey, K.; Leow, C.C.; Robbins, S.H. GITR ligand fusion protein agonist enhances the tumor antigen-specific CD8 T-cell response and leads to long-lasting memory. J. Immunother. Cancer 2017, 5, 47. [Google Scholar] [CrossRef]
- Piao, J.; Kamimura, Y.; Iwai, H.; Cao, Y.; Kikuchi, K.; Hashiguchi, M.; Masunaga, T.; Jiang, H.; Tamura, K.; Sakaguchi, S.; et al. Enhancement of T-cell-mediated anti-tumour immunity via the ectopically expressed glucocorticoid-induced tumour necrosis factor receptor-related receptor ligand (GITRL) on tumours. Immunology 2009, 127, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Kohm, A.P.; Williams, J.S.; Miller, S.D. Cutting edge: Ligation of the glucocorticoid-induced TNF receptor enhances autoreactive CD4+ T cell activation and experimental autoimmune encephalomyelitis. J. Immunol. (Baltim. Md. 1950) 2004, 172, 4686–4690. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, J.; Yamazaki, S.; Takahashi, T.; Ishida, Y.; Sakaguchi, S. Stimulation of CD25(+)CD4(+) regulatory T cells through GITR breaks immunological self-tolerance. Nat. Immunol. 2002, 3, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Knee, D.A.; Hewes, B.; Brogdon, J.L. Rationale for anti-GITR cancer immunotherapy. Eur. J. Cancer (Oxford Engl. 1990) 2016, 67, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.I.; McAdam, A.J.; Buhlmann, J.E.; Scott, S.; Lupher, M.L.; Greenfield, E.A.; Baum, P.R.; Fanslow, W.C.; Calderhead, D.M.; Freeman, G.J.; et al. Ox40-ligand has a critical costimulatory role in dendritic cell: T cell interactions. Immunity 1999, 11, 689–698. [Google Scholar] [CrossRef] [Green Version]
- Ishii, N.; Takahashi, T.; Soroosh, P.; Sugamura, K. OX40-OX40 ligand interaction in T-cell-mediated immunity and immunopathology. Adv. Immunol. 2010, 105, 63–98. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Ishii, N.; Takano, H.; Miura, S.; Ndhlovu, L.C.; Nose, M.; Noda, T.; Sugamura, K. Impairment of antigen-presenting cell function in mice lacking expression of OX40 ligand. J. Exp. Med. 2000, 191, 365–374. [Google Scholar] [CrossRef]
- Karulf, M.; Kelly, A.; Weinberg, A.D.; Gold, J.A. OX40 ligand regulates inflammation and mortality in the innate immune response to sepsis. J. Immunol. (Baltim. Md. 1950) 2010, 185, 4856–4862. [Google Scholar] [CrossRef] [Green Version]
- Linton, P.J.; Bautista, B.; Biederman, E.; Bradley, E.S.; Harbertson, J.; Kondrack, R.M.; Padrick, R.C.; Bradley, L.M. Costimulation via OX40L expressed by B cells is sufficient to determine the extent of primary CD4 cell expansion and Th2 cytokine secretion in vivo. J. Exp. Med. 2003, 197, 875–883. [Google Scholar] [CrossRef] [Green Version]
- Takeda, I.; Ine, S.; Killeen, N.; Ndhlovu, L.C.; Murata, K.; Satomi, S.; Sugamura, K.; Ishii, N. Distinct roles for the OX40-OX40 ligand interaction in regulatory and nonregulatory T cells. J. Immunol. (Baltim. Md. 1950) 2004, 172, 3580–3589. [Google Scholar] [CrossRef] [Green Version]
- Valzasina, B.; Guiducci, C.; Dislich, H.; Killeen, N.; Weinberg, A.D.; Colombo, M.P. Triggering of OX40 (CD134) on CD4(+)CD25+ T cells blocks their inhibitory activity: A novel regulatory role for OX40 and its comparison with GITR. Blood 2005, 105, 2845–2851. [Google Scholar] [CrossRef] [Green Version]
- De Smedt, T.; Smith, J.; Baum, P.; Fanslow, W.; Butz, E.; Maliszewski, C. Ox40 costimulation enhances the development of T cell responses induced by dendritic cells in vivo. J. Immunol. (Baltim. Md. 1950) 2002, 168, 661–670. [Google Scholar] [CrossRef]
- Flynn, S.; Toellner, K.M.; Raykundalia, C.; Goodall, M.; Lane, P. CD4 T cell cytokine differentiation: The B cell activation molecule, OX40 ligand, instructs CD4 T cells to express interleukin 4 and upregulates expression of the chemokine receptor, Blr-1. J. Exp. Med. 1998, 188, 297–304. [Google Scholar] [CrossRef]
- Higgins, L.M.; McDonald, S.A.; Whittle, N.; Crockett, N.; Shields, J.G.; MacDonald, T.T. Regulation of T cell activation in vitro and in vivo by targeting the OX40-OX40 ligand interaction: Amelioration of ongoing inflammatory bowel disease with an OX40-IgG fusion protein, but not with an OX40 ligand-IgG fusion protein. J. Immunol. (Baltim. Md. 1950) 1999, 162, 486–493. [Google Scholar]
- Jenkins, S.J.; Perona-Wright, G.; Worsley, A.G.; Ishii, N.; MacDonald, A.S. Dendritic cell expression of OX40 ligand acts as a costimulatory, not polarizing, signal for optimal Th2 priming and memory induction in vivo. J. Immunol. (Baltim. Md. 1950) 2007, 179, 3515–3523. [Google Scholar] [CrossRef]
- Ohshima, Y.; Yang, L.P.; Uchiyama, T.; Tanaka, Y.; Baum, P.; Sergerie, M.; Hermann, P.; Delespesse, G. OX40 costimulation enhances interleukin-4 (IL-4) expression at priming and promotes the differentiation of naive human CD4(+) T cells into high IL-4-producing effectors. Blood 1998, 92, 3338–3345. [Google Scholar] [CrossRef]
- Weinberg, A.D.; Rivera, M.M.; Prell, R.; Morris, A.; Ramstad, T.; Vetto, J.T.; Urba, W.J.; Alvord, G.; Bunce, C.; Shields, J. Engagement of the OX-40 receptor in vivo enhances antitumor immunity. J. Immunol. (Baltim. Md. 1950) 2000, 164, 2160–2169. [Google Scholar] [CrossRef]
- Rogers, P.R.; Song, J.; Gramaglia, I.; Killeen, N.; Croft, M. OX40 promotes Bcl-xL and Bcl-2 expression and is essential for long-term survival of CD4 T cells. Immunity 2001, 15, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Soroosh, P.; Ine, S.; Sugamura, K.; Ishii, N. OX40-OX40 ligand interaction through T cell-T cell contact contributes to CD4 T cell longevity. J. Immunol. (Baltim. Md. 1950) 2006, 176, 5975–5987. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Wang, Y.H.; Duramad, O.; Hanabuchi, S.; Perng, O.A.; Gilliet, M.; Qin, F.X.; Liu, Y.J. OX40 ligand shuts down IL-10-producing regulatory T cells. Proc. Natl. Acad. Sci. USA 2006, 103, 13138–13143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piconese, S.; Pittoni, P.; Burocchi, A.; Gorzanelli, A.; Care, A.; Tripodo, C.; Colombo, M.P. A non-redundant role for OX40 in the competitive fitness of Treg in response to IL-2. Eur. J. Immunol. 2010, 40, 2902–2913. [Google Scholar] [CrossRef] [PubMed]
- Vu, M.D.; Xiao, X.; Gao, W.; Degauque, N.; Chen, M.; Kroemer, A.; Killeen, N.; Ishii, N.; Li, X.C. OX40 costimulation turns off Foxp3+ Tregs. Blood 2007, 110, 2501–2510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.; Gong, W.; Demirci, G.; Liu, W.; Spoerl, S.; Chu, X.; Bishop, D.K.; Turka, L.A.; Li, X.C. New insights on OX40 in the control of T cell immunity and immune tolerance in vivo. J. Immunol. (Baltim. Md. 1950) 2012, 188, 892–901. [Google Scholar] [CrossRef] [Green Version]
- Buchan, S.; Manzo, T.; Flutter, B.; Rogel, A.; Edwards, N.; Zhang, L.; Sivakumaran, S.; Ghorashian, S.; Carpenter, B.; Bennett, C.; et al. OX40- and CD27-mediated costimulation synergizes with anti-PD-L1 blockade by forcing exhausted CD8+ T cells to exit quiescence. J. Immunol. (Baltim. Md. 1950) 2015, 194, 125–133. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Lin, Q.; Zhang, Z.; Zhang, L. Therapeutic strategies for the costimulatory molecule OX40 in T-cell-mediated immunity. Acta Pharm. Sin. B 2020, 10, 414–433. [Google Scholar] [CrossRef] [PubMed]
- Redmond, W.L.; Weinberg, A.D. Targeting OX40 and OX40L for the treatment of autoimmunity and cancer. Crit. Rev. Immunol. 2007, 27, 415–436. [Google Scholar] [CrossRef]
- Coussens, L.M.; Zitvogel, L.; Palucka, A.K. Neutralizing tumor-promoting chronic inflammation: A magic bullet? Science (N. Y.) 2013, 339, 286–291. [Google Scholar] [CrossRef] [Green Version]
- Hendriks, J.; Xiao, Y.; Rossen, J.W.; van der Sluijs, K.F.; Sugamura, K.; Ishii, N.; Borst, J. During viral infection of the respiratory tract, CD27, 4-1BB, and OX40 collectively determine formation of CD8+ memory T cells and their capacity for secondary expansion. J. Immunol. (Baltim. Md. 1950) 2005, 175, 1665–1676. [Google Scholar] [CrossRef] [Green Version]
- Bourgeois, C.; Rocha, B.; Tanchot, C. A role for CD40 expression on CD8+ T cells in the generation of CD8+ T cell memory. Science (N. Y.) 2002, 297, 2060–2063. [Google Scholar] [CrossRef]
- Tong, A.W.; Stone, M.J. Prospects for CD40-directed experimental therapy of human cancer. Cancer Gene Ther. 2003, 10, 1–13. [Google Scholar] [CrossRef]
- Bennett, S.R.; Carbone, F.R.; Karamalis, F.; Miller, J.F.; Heath, W.R. Induction of a CD8+ cytotoxic T lymphocyte response by cross-priming requires cognate CD4+ T cell help. J. Exp. Med. 1997, 186, 65–70. [Google Scholar] [CrossRef]
- Belz, G.T.; Wodarz, D.; Diaz, G.; Nowak, M.A.; Doherty, P.C. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. J. Virol. 2002, 76, 12388–12393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cella, M.; Scheidegger, D.; Palmer-Lehmann, K.; Lane, P.; Lanzavecchia, A.; Alber, G. Ligation of CD40 on dendritic cells triggers production of high levels of interleukin-12 and enhances T cell stimulatory capacity: T-T help via APC activation. J. Exp. Med. 1996, 184, 747–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diehl, L.; den Boer, A.T.; Schoenberger, S.P.; van der Voort, E.I.; Schumacher, T.N.; Melief, C.J.; Offringa, R.; Toes, R.E. CD40 activation in vivo overcomes peptide-induced peripheral cytotoxic T-lymphocyte tolerance and augments anti-tumor vaccine efficacy. Nat. Med. 1999, 5, 774–779. [Google Scholar] [CrossRef]
- Kedl, R.M.; Jordan, M.; Potter, T.; Kappler, J.; Marrack, P.; Dow, S. CD40 stimulation accelerates deletion of tumor-specific CD8(+) T cells in the absence of tumor-antigen vaccination. Proc. Natl. Acad. Sci. USA 2001, 98, 10811–10816. [Google Scholar] [CrossRef] [Green Version]
- Ahrends, T.; Busselaar, J.; Severson, T.M.; Babala, N.; de Vries, E.; Bovens, A.; Wessels, L.; van Leeuwen, F.; Borst, J. CD4(+) T cell help creates memory CD8(+) T cells with innate and help-independent recall capacities. Nat. Commun. 2019, 10, 5531. [Google Scholar] [CrossRef]
- Vonderheide, R.H.; Glennie, M.J. Agonistic CD40 antibodies and cancer therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 1035–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, A.M.; Schildknecht, A.; Xiao, Y.; van den Broek, M.; Borst, J. Expression of costimulatory ligand CD70 on steady-state dendritic cells breaks CD8+ T cell tolerance and permits effective immunity. Immunity 2008, 29, 934–946. [Google Scholar] [CrossRef] [Green Version]
- Bullock, T.N.; Yagita, H. Induction of CD70 on dendritic cells through CD40 or TLR stimulation contributes to the development of CD8+ T cell responses in the absence of CD4+ T cells. J. Immunol. (Baltim. Md. 1950) 2005, 174, 710–717. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, P.J.; McWilliams, J.A.; Haluszczak, C.; Yagita, H.; Kedl, R.M. Combined TLR/CD40 stimulation mediates potent cellular immunity by regulating dendritic cell expression of CD70 in vivo. J. Immunol. (Baltim. Md. 1950) 2007, 178, 1564–1572. [Google Scholar] [CrossRef] [Green Version]
- Feau, S.; Garcia, Z.; Arens, R.; Yagita, H.; Borst, J.; Schoenberger, S.P. The CD4(+) T-cell help signal is transmitted from APC to CD8(+) T-cells via CD27-CD70 interactions. Nat. Commun. 2012, 3, 948. [Google Scholar] [CrossRef]
- French, R.R.; Taraban, V.Y.; Crowther, G.R.; Rowley, T.F.; Gray, J.C.; Johnson, P.W.; Tutt, A.L.; Al-Shamkhani, A.; Glennie, M.J. Eradication of lymphoma by CD8 T cells following anti-CD40 monoclonal antibody therapy is critically dependent on CD27 costimulation. Blood 2007, 109, 4810–4815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arens, R.; Schepers, K.; Nolte, M.A.; van Oosterwijk, M.F.; van Lier, R.A.; Schumacher, T.N.; van Oers, M.H. Tumor rejection induced by CD70-mediated quantitative and qualitative effects on effector CD8+ T cell formation. J. Exp. Med. 2004, 199, 1595–1605. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, J.; Gravestein, L.A.; Tesselaar, K.; van Lier, R.A.; Schumacher, T.N.; Borst, J. CD27 is required for generation and long-term maintenance of T cell immunity. Nat. Immunol. 2000, 1, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Kuka, M.; Munitic, I.; Giardino Torchia, M.L.; Ashwell, J.D. CD70 is downregulated by interaction with CD27. J. Immunol. (Baltim. Md. 1950) 2013, 191, 2282–2289. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, P.J.; Kedl, R.M. An alternative signal 3: CD8(+) T cell memory independent of IL-12 and type I IFN is dependent on CD27/OX40 signaling. Vaccine 2012, 30, 1154–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares, H.; Waechter, H.; Glaichenhaus, N.; Mougneau, E.; Yagita, H.; Mizenina, O.; Dudziak, D.; Nussenzweig, M.C.; Steinman, R.M. A subset of dendritic cells induces CD4+ T cells to produce IFN-gamma by an IL-12-independent but CD70-dependent mechanism in vivo. J. Exp. Med. 2007, 204, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Tesselaar, K.; Arens, R.; van Schijndel, G.M.; Baars, P.A.; van der Valk, M.A.; Borst, J.; van Oers, M.H.; van Lier, R.A. Lethal T cell immunodeficiency induced by chronic costimulation via CD27-CD70 interactions. Nat. Immunol. 2003, 4, 49–54. [Google Scholar] [CrossRef]
- Van de Ven, K.; Borst, J. Targeting the T-cell co-stimulatory CD27/CD70 pathway in cancer immunotherapy: Rationale and potential. Immunotherapy 2015, 7, 655–667. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.Z.; Grote, D.M.; Xiu, B.; Ziesmer, S.C.; Price-Troska, T.L.; Hodge, L.S.; Yates, D.M.; Novak, A.J.; Ansell, S.M. TGF-beta upregulates CD70 expression and induces exhaustion of effector memory T cells in B-cell non-Hodgkin’s lymphoma. Leukemia 2014, 28, 1872–1884. [Google Scholar] [CrossRef] [Green Version]
- Buchan, S.L.; Fallatah, M.; Thirdborough, S.M.; Taraban, V.Y.; Rogel, A.; Thomas, L.J.; Penfold, C.A.; He, L.Z.; Curran, M.A.; Keler, T.; et al. PD-1 Blockade and CD27 Stimulation Activate Distinct Transcriptional Programs That Synergize for CD8(+) T-Cell-Driven Antitumor Immunity. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 2383–2394. [Google Scholar] [CrossRef] [Green Version]
- Agata, Y.; Kawasaki, A.; Nishimura, H.; Ishida, Y.; Tsubata, T.; Yagita, H.; Honjo, T. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int. Immunol. 1996, 8, 765–772. [Google Scholar] [CrossRef] [Green Version]
- Liang, S.C.; Latchman, Y.E.; Buhlmann, J.E.; Tomczak, M.F.; Horwitz, B.H.; Freeman, G.J.; Sharpe, A.H. Regulation of PD-1, PD-L1, and PD-L2 expression during normal and autoimmune responses. Eur. J. Immunol. 2003, 33, 2706–2716. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, A.H.; Pauken, K.E. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 2018, 18, 153–167. [Google Scholar] [CrossRef]
- Hui, E.; Cheung, J.; Zhu, J.; Su, X.; Taylor, M.J.; Wallweber, H.A.; Sasmal, D.K.; Huang, J.; Kim, J.M.; Mellman, I.; et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science (N. Y.) 2017, 355, 1428–1433. [Google Scholar] [CrossRef] [PubMed]
- Parry, R.V.; Chemnitz, J.M.; Frauwirth, K.A.; Lanfranco, A.R.; Braunstein, I.; Kobayashi, S.V.; Linsley, P.S.; Thompson, C.B.; Riley, J.L. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell. Boil. 2005, 25, 9543–9553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokosuka, T.; Takamatsu, M.; Kobayashi-Imanishi, W.; Hashimoto-Tane, A.; Azuma, M.; Saito, T. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med. 2012, 209, 1201–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barber, D.L.; Wherry, E.J.; Masopust, D.; Zhu, B.; Allison, J.P.; Sharpe, A.H.; Freeman, G.J.; Ahmed, R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006, 439, 682–687. [Google Scholar] [CrossRef]
- Crawford, A.; Angelosanto, J.M.; Kao, C.; Doering, T.A.; Odorizzi, P.M.; Barnett, B.E.; Wherry, E.J. Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity 2014, 40, 289–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauken, K.E.; Wherry, E.J. Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 2015, 36, 265–276. [Google Scholar] [CrossRef] [Green Version]
- Kamphorst, A.O.; Wieland, A.; Nasti, T.; Yang, S.; Zhang, R.; Barber, D.L.; Konieczny, B.T.; Daugherty, C.Z.; Koenig, L.; Yu, K.; et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science (N. Y.) 2017, 355, 1423–1427. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.H.; Qiu, J.; O’Sullivan, D.; Buck, M.D.; Noguchi, T.; Curtis, J.D.; Chen, Q.; Gindin, M.; Gubin, M.M.; van der Windt, G.J.; et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 2015, 162, 1229–1241. [Google Scholar] [CrossRef] [Green Version]
- O’Sullivan, D.; Pearce, E.L. Targeting T cell metabolism for therapy. Trends Immunol. 2015, 36, 71–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patsoukis, N.; Bardhan, K.; Chatterjee, P.; Sari, D.; Liu, B.; Bell, L.N.; Karoly, E.D.; Freeman, G.J.; Petkova, V.; Seth, P.; et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 2015, 6, 6692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, A.; Harker, J.A.; Chanthong, K.; Stevenson, P.G.; Zuniga, E.I.; Rudd, C.E. Glycogen Synthase Kinase 3 Inactivation Drives T-bet-Mediated Downregulation of Co-receptor PD-1 to Enhance CD8(+) Cytolytic T Cell Responses. Immunity 2016, 44, 274–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, H.; Zhu, G.; Tamada, K.; Chen, L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 1999, 5, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Freeman, G.J.; Long, A.J.; Iwai, Y.; Bourque, K.; Chernova, T.; Nishimura, H.; Fitz, L.J.; Malenkovich, N.; Okazaki, T.; Byrne, M.C.; et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 2000, 192, 1027–1034. [Google Scholar] [CrossRef] [Green Version]
- Latchman, Y.; Wood, C.R.; Chernova, T.; Chaudhary, D.; Borde, M.; Chernova, I.; Iwai, Y.; Long, A.J.; Brown, J.A.; Nunes, R.; et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2001, 2, 261–268. [Google Scholar] [CrossRef]
- Tseng, S.Y.; Otsuji, M.; Gorski, K.; Huang, X.; Slansky, J.E.; Pai, S.I.; Shalabi, A.; Shin, T.; Pardoll, D.M.; Tsuchiya, H. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J. Exp. Med. 2001, 193, 839–846. [Google Scholar] [CrossRef]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [Green Version]
- Juneja, V.R.; McGuire, K.A.; Manguso, R.T.; LaFleur, M.W.; Collins, N.; Haining, W.N.; Freeman, G.J.; Sharpe, A.H. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J. Exp. Med. 2017, 214, 895–904. [Google Scholar] [CrossRef]
- Eppihimer, M.J.; Gunn, J.; Freeman, G.J.; Greenfield, E.A.; Chernova, T.; Erickson, J.; Leonard, J.P. Expression and regulation of the PD-L1 immunoinhibitory molecule on microvascular endothelial cells. Microcirculation (New York 1994) 2002, 9, 133–145. [Google Scholar] [CrossRef]
- Askenase, M.H.; Han, S.J.; Byrd, A.L.; Morais da Fonseca, D.; Bouladoux, N.; Wilhelm, C.; Konkel, J.E.; Hand, T.W.; Lacerda-Queiroz, N.; Su, X.Z.; et al. Bone-Marrow-Resident NK Cells Prime Monocytes for Regulatory Function during Infection. Immunity 2015, 42, 1130–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, H.; Strome, S.E.; Salomao, D.R.; Tamura, H.; Hirano, F.; Flies, D.B.; Roche, P.C.; Lu, J.; Zhu, G.; Tamada, K.; et al. Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 2002, 8, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Berger, R.; Rotem-Yehudar, R.; Slama, G.; Landes, S.; Kneller, A.; Leiba, M.; Koren-Michowitz, M.; Shimoni, A.; Nagler, A. Phase I safety and pharmacokinetic study of CT-011, a humanized antibody interacting with PD-1, in patients with advanced hematologic malignancies. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 3044–3051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brahmer, J.R.; Tykodi, S.S.; Chow, L.Q.; Hwu, W.J.; Topalian, S.L.; Hwu, P.; Drake, C.G.; Camacho, L.H.; Kauh, J.; Odunsi, K.; et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 2012, 366, 2455–2465. [Google Scholar] [CrossRef] [Green Version]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef]
- Wolchok, J.D.; Kluger, H.; Callahan, M.K.; Postow, M.A.; Rizvi, N.A.; Lesokhin, A.M.; Segal, N.H.; Ariyan, C.E.; Gordon, R.A.; Reed, K.; et al. Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J. Med. 2013, 369, 122–133. [Google Scholar] [CrossRef] [Green Version]
- Butte, M.J.; Keir, M.E.; Phamduy, T.B.; Sharpe, A.H.; Freeman, G.J. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity 2007, 27, 111–122. [Google Scholar] [CrossRef] [Green Version]
- Francisco, L.M.; Salinas, V.H.; Brown, K.E.; Vanguri, V.K.; Freeman, G.J.; Kuchroo, V.K.; Sharpe, A.H. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J. Exp. Med. 2009, 206, 3015–3029. [Google Scholar] [CrossRef]
- Wang, W.; Lau, R.; Yu, D.; Zhu, W.; Korman, A.; Weber, J. PD1 blockade reverses the suppression of melanoma antigen-specific CTL by CD4+ CD25(Hi) regulatory T cells. Int. Immunol. 2009, 21, 1065–1077. [Google Scholar] [CrossRef] [Green Version]
- Trefny, M.P.; Kaiser, M.; Stanczak, M.A.; Herzig, P.; Savic, S.; Wiese, M.; Lardinois, D.; Laubli, H.; Uhlenbrock, F.; Zippelius, A. PD-1(+) natural killer cells in human non-small cell lung cancer can be activated by PD-1/PD-L1 blockade. Cancer Immunol. Immunother. 2020. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Ohashi, P.S. Clinical blockade of PD1 and LAG3--potential mechanisms of action. Nat. Rev. Immunol. 2015, 15, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, H.; Muskens, F.; Willart, M.; Hijdra, D.; van Assema, F.B.; Coyle, A.J.; Hoogsteden, H.C.; Lambrecht, B.N. Contribution of the PD-1 ligands/PD-1 signaling pathway to dendritic cell-mediated CD4+ T cell activation. Eur. J. Immunol. 2006, 36, 2472–2482. [Google Scholar] [CrossRef] [PubMed]
- Diskin, B.; Adam, S.; Cassini, M.F.; Sanchez, G.; Liria, M.; Aykut, B.; Buttar, C.; Li, E.; Sundberg, B.; Salas, R.D.; et al. PD-L1 engagement on T cells promotes self-tolerance and suppression of neighboring macrophages and effector T cells in cancer. Nat. Immunol. 2020, 21, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Lines, J.L.; Pantazi, E.; Mak, J.; Sempere, L.F.; Wang, L.; O’Connell, S.; Ceeraz, S.; Suriawinata, A.A.; Yan, S.; Ernstoff, M.S.; et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014, 74, 1924–1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenbaum, S.R.; Knecht, M.; Mollaee, M.; Zhong, Z.; Erkes, D.A.; McCue, P.A.; Chervoneva, I.; Berger, A.C.; Lo, J.A.; Fisher, D.E.; et al. FOXD3 Regulates VISTA Expression in Melanoma. Cell Rep. 2020, 30, 510–524 e516. [Google Scholar] [CrossRef]
- Yoon, K.W.; Byun, S.; Kwon, E.; Hwang, S.Y.; Chu, K.; Hiraki, M.; Jo, S.H.; Weins, A.; Hakroush, S.; Cebulla, A.; et al. Control of signaling-mediated clearance of apoptotic cells by the tumor suppressor p53. Science (N. Y.) 2015, 349, 1261669. [Google Scholar] [CrossRef] [Green Version]
- Nowak, E.C.; Lines, J.L.; Varn, F.S.; Deng, J.; Sarde, A.; Mabaera, R.; Kuta, A.; Le Mercier, I.; Cheng, C.; Noelle, R.J. Immunoregulatory functions of VISTA. Immunol. Rev. 2017, 276, 66–79. [Google Scholar] [CrossRef]
- Liu, J.; Yuan, Y.; Chen, W.; Putra, J.; Suriawinata, A.A.; Schenk, A.D.; Miller, H.E.; Guleria, I.; Barth, R.J.; Huang, Y.H.; et al. Immune-checkpoint proteins VISTA and PD-1 nonredundantly regulate murine T-cell responses. Proc. Natl. Acad. Sci. USA 2015, 112, 6682–6687. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Rubinstein, R.; Lines, J.L.; Wasiuk, A.; Ahonen, C.; Guo, Y.; Lu, L.F.; Gondek, D.; Wang, Y.; Fava, R.A.; et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J. Exp. Med. 2011, 208, 577–592. [Google Scholar] [CrossRef]
- Flies, D.B.; Han, X.; Higuchi, T.; Zheng, L.; Sun, J.; Ye, J.J.; Chen, L. Coinhibitory receptor PD-1H preferentially suppresses CD4(+) T cell-mediated immunity. J. Clin. Investig. 2014, 124, 1966–1975. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Xu, W.; Yuan, Y.; Ayithan, N.; Imai, Y.; Wu, X.; Miller, H.; Olson, M.; Feng, Y.; Huang, Y.H.; et al. Immune-checkpoint protein VISTA critically regulates the IL-23/IL-17 inflammatory axis. Sci, Rep. 2017, 7, 1485. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Le Mercier, I.; Putra, J.; Chen, W.; Liu, J.; Schenk, A.D.; Nowak, E.C.; Suriawinata, A.A.; Li, J.; Noelle, R.J. Disruption of the immune-checkpoint VISTA gene imparts a proinflammatory phenotype with predisposition to the development of autoimmunity. Proc. Natl. Acad. Sci. USA 2014, 111, 14846–14851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, Y.; Ohno, T.; Nishii, N.; Harada, K.; Yagita, H.; Azuma, M. Differential contribution of three immune checkpoint (VISTA, CTLA-4, PD-1) pathways to antitumor responses against squamous cell carcinoma. Oral Oncol. 2016, 57, 54–60. [Google Scholar] [CrossRef]
- Le Mercier, I.; Chen, W.; Lines, J.L.; Day, M.; Li, J.; Sergent, P.; Noelle, R.J.; Wang, L. VISTA Regulates the Development of Protective Antitumor Immunity. Cancer Res. 2014, 74, 1933–1944. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Ward, J.F.; Pettaway, C.A.; Shi, L.Z.; Subudhi, S.K.; Vence, L.M.; Zhao, H.; Chen, J.; Chen, H.; Efstathiou, E.; et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat. Med. 2017, 23, 551–555. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Flies, D.B. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 2013, 13, 227–242. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell 2015, 161, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science (N. Y.) 2015, 348, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Coombes, J.L.; Siddiqui, K.R.; Arancibia-Carcamo, C.V.; Hall, J.; Sun, C.M.; Belkaid, Y.; Powrie, F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 2007, 204, 1757–1764. [Google Scholar] [CrossRef]
- Sun, C.M.; Hall, J.A.; Blank, R.B.; Bouladoux, N.; Oukka, M.; Mora, J.R.; Belkaid, Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 2007, 204, 1775–1785. [Google Scholar] [CrossRef] [Green Version]
- Alexandre, Y.O.; Ghilas, S.; Sanchez, C.; Le Bon, A.; Crozat, K.; Dalod, M. XCR1+ dendritic cells promote memory CD8+ T cell recall upon secondary infections with Listeria monocytogenes or certain viruses. J. Exp. Med. 2016, 213, 75–92. [Google Scholar] [CrossRef] [Green Version]
- Draheim, M.; Wlodarczyk, M.F.; Crozat, K.; Saliou, J.M.; Alayi, T.D.; Tomavo, S.; Hassan, A.; Salvioni, A.; Demarta-Gatsi, C.; Sidney, J.; et al. Profiling MHC II immunopeptidome of blood-stage malaria reveals that cDC1 control the functionality of parasite-specific CD4 T cells. EMBO Mol. Med. 2017, 9, 1605–1621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrand, K.J.; Dickgreber, N.; Stoitzner, P.; Ronchese, F.; Petersen, T.R.; Hermans, I.F. Langerin+ CD8alpha+ dendritic cells are critical for cross-priming and IL-12 production in response to systemic antigens. J. Immunol. (Baltim. Md. 1950) 2009, 183, 7732–7742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, C.J.; Ornelles, D.A.; Mitchell, L.M.; Brzoza-Lewis, K.L.; Hiltbold, E.M. IL-12 produced by dendritic cells augments CD8+ T cell activation through the production of the chemokines CCL1 and CCL17. J. Immunol. (Baltim. Md. 1950) 2008, 181, 8576–8584. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.H.; Fan, Y.T.; Dias, A.; Esper, L.; Corn, R.A.; Bafica, A.; Machado, F.S.; Aliberti, J. Cutting edge: Dendritic cells are essential for in vivo IL-12 production and development of resistance against Toxoplasma gondii infection in mice. J. Immunol. (Baltim. Md. 1950) 2006, 177, 31–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado-Lopez, R.; De Smedt, T.; Michel, P.; Godfroid, J.; Pajak, B.; Heirman, C.; Thielemans, K.; Leo, O.; Urbain, J.; Moser, M. CD8alpha+ and CD8alpha- subclasses of dendritic cells direct the development of distinct T helper cells in vivo. J. Exp. Med. 1999, 189, 587–592. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Lopez, M.; Iborra, S.; Conde-Garrosa, R.; Sancho, D. Batf3-dependent CD103+ dendritic cells are major producers of IL-12 that drive local Th1 immunity against Leishmania major infection in mice. Eur. J. Immunol. 2015, 45, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Mashayekhi, M.; Sandau, M.M.; Dunay, I.R.; Frickel, E.M.; Khan, A.; Goldszmid, R.S.; Sher, A.; Ploegh, H.L.; Murphy, T.L.; Sibley, L.D.; et al. CD8alpha(+) dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity 2011, 35, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Persky, M.E.; Murphy, K.M.; Farrar, J.D. IL-12, but not IFN-alpha, promotes STAT4 activation and Th1 development in murine CD4+ T cells expressing a chimeric murine/human Stat2 gene. J. Immunol. (Baltim. Md. 1950) 2005, 174, 294–301. [Google Scholar] [CrossRef] [Green Version]
- Pulendran, B.; Smith, J.L.; Caspary, G.; Brasel, K.; Pettit, D.; Maraskovsky, E.; Maliszewski, C.R. Distinct dendritic cell subsets differentially regulate the class of immune response in vivo. Proc. Natl. Acad. Sci. USA 1999, 96, 1036–1041. [Google Scholar] [CrossRef] [Green Version]
- Reis e Sousa, C.; Hieny, S.; Scharton-Kersten, T.; Jankovic, D.; Charest, H.; Germain, R.N.; Sher, A. In vivo microbial stimulation induces rapid CD40 ligand-independent production of interleukin 12 by dendritic cells and their redistribution to T cell areas. J. Exp. Med. 1997, 186, 1819–1829. [Google Scholar] [CrossRef] [Green Version]
- Kretschmer, K.; Apostolou, I.; Hawiger, D.; Khazaie, K.; Nussenzweig, M.C.; von Boehmer, H. Inducing and expanding regulatory T cell populations by foreign antigen. Nat. Immunol. 2005, 6, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.A.; Lu, Y.B.; Wang, W.D.; Liu, G.B.; Chen, C.; Shen, L.; Luo, H.L.; Xu, H.; Peng, Y.; Luo, H.; et al. BTLA-Expressing Dendritic Cells in Patients With Tuberculosis Exhibit Reduced Production of IL-12/IFN-alpha and Increased Production of IL-4 and TGF-beta, Favoring Th2 and Foxp3(+) Treg Polarization. Front. Immunol. 2020, 11, 518. [Google Scholar] [CrossRef]
- Joffre, O.P.; Segura, E.; Savina, A.; Amigorena, S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012, 12, 557–569. [Google Scholar] [CrossRef]
- Luckashenak, N.; Schroeder, S.; Endt, K.; Schmidt, D.; Mahnke, K.; Bachmann, M.F.; Marconi, P.; Deeg, C.A.; Brocker, T. Constitutive crosspresentation of tissue antigens by dendritic cells controls CD8+ T cell tolerance in vivo. Immunity 2008, 28, 521–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair-Gupta, P.; Blander, J.M. An updated view of the intracellular mechanisms regulating cross-presentation. Front. Immunol. 2013, 4, 401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dillon, S.; Agrawal, A.; Van Dyke, T.; Landreth, G.; McCauley, L.; Koh, A.; Maliszewski, C.; Akira, S.; Pulendran, B. A Toll-like receptor 2 ligand stimulates Th2 responses in vivo, via induction of extracellular signal-regulated kinase mitogen-activated protein kinase and c-Fos in dendritic cells. J. Immunol. (Baltim. Md. 1950) 2004, 172, 4733–4743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitajima, M.; Ziegler, S.F. Cutting edge: Identification of the thymic stromal lymphopoietin-responsive dendritic cell subset critical for initiation of type 2 contact hypersensitivity. J. Immunol. (Baltim. Md. 1950) 2013, 191, 4903–4907. [Google Scholar] [CrossRef] [Green Version]
- Bettelli, E.; Carrier, Y.; Gao, W.; Korn, T.; Strom, T.B.; Oukka, M.; Weiner, H.L.; Kuchroo, V.K. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 2006, 441, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; McKenzie, B.S.; Zhou, L.; Tadokoro, C.E.; Lepelley, A.; Lafaille, J.J.; Cua, D.J.; Littman, D.R. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 2006, 126, 1121–1133. [Google Scholar] [CrossRef] [Green Version]
- Mangan, P.R.; Harrington, L.E.; O’Quinn, D.B.; Helms, W.S.; Bullard, D.C.; Elson, C.O.; Hatton, R.D.; Wahl, S.M.; Schoeb, T.R.; Weaver, C.T. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 2006, 441, 231–234. [Google Scholar] [CrossRef]
- Nurieva, R.; Yang, X.O.; Martinez, G.; Zhang, Y.; Panopoulos, A.D.; Ma, L.; Schluns, K.; Tian, Q.; Watowich, S.S.; Jetten, A.M.; et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 2007, 448, 480–483. [Google Scholar] [CrossRef]
- Veldhoen, M.; Hocking, R.J.; Atkins, C.J.; Locksley, R.M.; Stockinger, B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 2006, 24, 179–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Danilenko, D.M.; Valdez, P.; Kasman, I.; Eastham-Anderson, J.; Wu, J.; Ouyang, W. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature 2007, 445, 648–651. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ivanov, I.I.; Spolski, R.; Min, R.; Shenderov, K.; Egawa, T.; Levy, D.E.; Leonard, W.J.; Littman, D.R. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 2007, 8, 967–974. [Google Scholar] [CrossRef] [PubMed]
- Korn, T.; Bettelli, E.; Gao, W.; Awasthi, A.; Jager, A.; Strom, T.B.; Oukka, M.; Kuchroo, V.K. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature 2007, 448, 484–487. [Google Scholar] [CrossRef]
- Korn, T.; Mitsdoerffer, M.; Croxford, A.L.; Awasthi, A.; Dardalhon, V.A.; Galileos, G.; Vollmar, P.; Stritesky, G.L.; Kaplan, M.H.; Waisman, A.; et al. IL-6 controls Th17 immunity in vivo by inhibiting the conversion of conventional T cells into Foxp3+ regulatory T cells. Proc. Natl. Acad. Sci. USA 2008, 105, 18460–18465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briseno, C.G.; Satpathy, A.T.; Davidson, J.T.t.; Ferris, S.T.; Durai, V.; Bagadia, P.; O’Connor, K.W.; Theisen, D.J.; Murphy, T.L.; Murphy, K.M. Notch2-dependent DC2s mediate splenic germinal center responses. Proc. Natl. Acad. Sci. USA 2018, 115, 10726–10731. [Google Scholar] [CrossRef] [Green Version]
- Murphy, K.M.; Reiner, S.L. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2002, 2, 933–944. [Google Scholar] [CrossRef]
- Paul, W.E.; Zhu, J. How are T(H)2-type immune responses initiated and amplified? Nat. Rev. Immunol. 2010, 10, 225–235. [Google Scholar] [CrossRef]
- Proietto, A.I.; van Dommelen, S.; Zhou, P.; Rizzitelli, A.; D’Amico, A.; Steptoe, R.J.; Naik, S.H.; Lahoud, M.H.; Liu, Y.; Zheng, P.; et al. Dendritic cells in the thymus contribute to T-regulatory cell induction. Proc. Natl. Acad. Sci. USA 2008, 105, 19869–19874. [Google Scholar] [CrossRef] [Green Version]
- Cohn, L.; Chatterjee, B.; Esselborn, F.; Smed-Sorensen, A.; Nakamura, N.; Chalouni, C.; Lee, B.C.; Vandlen, R.; Keler, T.; Lauer, P.; et al. Antigen delivery to early endosomes eliminates the superiority of human blood BDCA3+ dendritic cells at cross presentation. J. Exp. Med. 2013, 210, 1049–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalod, M.; Hamilton, T.; Salomon, R.; Salazar-Mather, T.P.; Henry, S.C.; Hamilton, J.D.; Biron, C.A. Dendritic cell responses to early murine cytomegalovirus infection: Subset functional specialization and differential regulation by interferon alpha/beta. J. Exp. Med. 2003, 197, 885–898. [Google Scholar] [CrossRef] [Green Version]
- Manetti, R.; Parronchi, P.; Giudizi, M.G.; Piccinni, M.P.; Maggi, E.; Trinchieri, G.; Romagnani, S. Natural killer cell stimulatory factor (interleukin 12 [IL-12]) induces T helper type 1 (Th1)-specific immune responses and inhibits the development of IL-4-producing Th cells. J. Exp. Med. 1993, 177, 1199–1204. [Google Scholar] [CrossRef] [Green Version]
- Mittag, D.; Proietto, A.I.; Loudovaris, T.; Mannering, S.I.; Vremec, D.; Shortman, K.; Wu, L.; Harrison, L.C. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J. Immunol. (Baltim. Md. 1950) 2011, 186, 6207–6217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Napolitani, G.; Rinaldi, A.; Bertoni, F.; Sallusto, F.; Lanzavecchia, A. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 2005, 6, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Nizzoli, G.; Krietsch, J.; Weick, A.; Steinfelder, S.; Facciotti, F.; Gruarin, P.; Bianco, A.; Steckel, B.; Moro, M.; Crosti, M.; et al. Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood 2013, 122, 932–942. [Google Scholar] [CrossRef] [Green Version]
- Piccioli, D.; Tavarini, S.; Borgogni, E.; Steri, V.; Nuti, S.; Sammicheli, C.; Bardelli, M.; Montagna, D.; Locatelli, F.; Wack, A. Functional specialization of human circulating CD16 and CD1c myeloid dendritic-cell subsets. Blood 2007, 109, 5371–5379. [Google Scholar] [CrossRef] [Green Version]
- Poulin, L.F.; Salio, M.; Griessinger, E.; Anjos-Afonso, F.; Craciun, L.; Chen, J.L.; Keller, A.M.; Joffre, O.; Zelenay, S.; Nye, E.; et al. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J. Exp. Med. 2010, 207, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Segura, E.; Durand, M.; Amigorena, S. Similar antigen cross-presentation capacity and phagocytic functions in all freshly isolated human lymphoid organ-resident dendritic cells. J. Exp. Med. 2013, 210, 1035–1047. [Google Scholar] [CrossRef] [Green Version]
- Lauterbach, H.; Bathke, B.; Gilles, S.; Traidl-Hoffmann, C.; Luber, C.A.; Fejer, G.; Freudenberg, M.A.; Davey, G.M.; Vremec, D.; Kallies, A.; et al. Mouse CD8alpha+ DCs and human BDCA3+ DCs are major producers of IFN-lambda in response to poly IC. J. Exp. Med. 2010, 207, 2703–2717. [Google Scholar] [CrossRef]
- Olsen, I.; Sollid, L.M. Pitfalls in determining the cytokine profile of human T cells. J. Immunol. Methods 2013, 390, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.; Benson, A.; Kuzmich, L.; DeFranco, A.L.; Yarovinsky, F. Critical coordination of innate immune defense against Toxoplasma gondii by dendritic cells responding via their Toll-like receptors. Proc. Natl. Acad. Sci. USA 2011, 108, 278–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bottcher, J.P.; Bonavita, E.; Chakravarty, P.; Blees, H.; Cabeza-Cabrerizo, M.; Sammicheli, S.; Rogers, N.C.; Sahai, E.; Zelenay, S.; Reis, E.S.C. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 2018, 172, 1022–1037.e1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagadia, P.; Huang, X.; Liu, T.T.; Murphy, K.M. Shared Transcriptional Control of Innate Lymphoid Cell and Dendritic Cell Development. Annu. Rev. Cell Dev. Boil. 2019, 35, 381–406. [Google Scholar] [CrossRef] [PubMed]
- Robinette, M.L.; Colonna, M. Immune modules shared by innate lymphoid cells and T cells. J. Allergy Clin. Immunol. 2016, 138, 1243–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diebold, S.S.; Montoya, M.; Unger, H.; Alexopoulou, L.; Roy, P.; Haswell, L.E.; Al-Shamkhani, A.; Flavell, R.; Borrow, P.; Reis e Sousa, C. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature 2003, 424, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Uematsu, S.; Matsui, K.; Tsujimura, T.; Takeda, K.; Fujita, T.; Takeuchi, O.; et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity 2005, 23, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Salazar, F.; Awuah, D.; Negm, O.H.; Shakib, F.; Ghaemmaghami, A.M. The role of indoleamine 2,3-dioxygenase-aryl hydrocarbon receptor pathway in the TLR4-induced tolerogenic phenotype in human DCs. Sci. Rep. 2017, 7, 43337. [Google Scholar] [CrossRef]
- Tailor, P.; Tamura, T.; Ozato, K. IRF family proteins and type I interferon induction in dendritic cells. Cell Res. 2006, 16, 134–140. [Google Scholar] [CrossRef] [Green Version]
- Tailor, P.; Tamura, T.; Kong, H.J.; Kubota, T.; Kubota, M.; Borghi, P.; Gabriele, L.; Ozato, K. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity 2007, 27, 228–239. [Google Scholar] [CrossRef] [Green Version]
- Fuertes, M.B.; Kacha, A.K.; Kline, J.; Woo, S.R.; Kranz, D.M.; Murphy, K.M.; Gajewski, T.F. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J. Exp. Med. 2011, 208, 2005–2016. [Google Scholar] [CrossRef] [Green Version]
- Ito, T.; Amakawa, R.; Inaba, M.; Ikehara, S.; Inaba, K.; Fukuhara, S. Differential regulation of human blood dendritic cell subsets by IFNs. J. Immunol. (Baltim. Md. 1950) 2001, 166, 2961–2969. [Google Scholar] [CrossRef] [PubMed]
- Le Bon, A.; Etchart, N.; Rossmann, C.; Ashton, M.; Hou, S.; Gewert, D.; Borrow, P.; Tough, D.F. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat. Immunol. 2003, 4, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, S.; Mattei, F.; Sistigu, A.; Bracci, L.; Spadaro, F.; Sanchez, M.; Spada, M.; Belardelli, F.; Gabriele, L.; Schiavoni, G. Type I IFNs control antigen retention and survival of CD8alpha(+) dendritic cells after uptake of tumor apoptotic cells leading to cross-priming. J. Immunol. (Baltim. Md. 1950) 2011, 186, 5142–5150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parlato, S.; Romagnoli, G.; Spadaro, F.; Canini, I.; Sirabella, P.; Borghi, P.; Ramoni, C.; Filesi, I.; Biocca, S.; Gabriele, L.; et al. LOX-1 as a natural IFN-alpha-mediated signal for apoptotic cell uptake and antigen presentation in dendritic cells. Blood 2010, 115, 1554–1563. [Google Scholar] [CrossRef] [Green Version]
- Schiavoni, G.; Sistigu, A.; Valentini, M.; Mattei, F.; Sestili, P.; Spadaro, F.; Sanchez, M.; Lorenzi, S.; D’Urso, M.T.; Belardelli, F.; et al. Cyclophosphamide synergizes with type I interferons through systemic dendritic cell reactivation and induction of immunogenic tumor apoptosis. Cancer Res. 2011, 71, 768–778. [Google Scholar] [CrossRef] [Green Version]
- Spadaro, F.; Lapenta, C.; Donati, S.; Abalsamo, L.; Barnaba, V.; Belardelli, F.; Santini, S.M.; Ferrantini, M. IFN-alpha enhances cross-presentation in human dendritic cells by modulating antigen survival, endocytic routing, and processing. Blood 2012, 119, 1407–1417. [Google Scholar] [CrossRef] [Green Version]
- Diamond, M.S.; Kinder, M.; Matsushita, H.; Mashayekhi, M.; Dunn, G.P.; Archambault, J.M.; Lee, H.; Arthur, C.D.; White, J.M.; Kalinke, U.; et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 2011, 208, 1989–2003. [Google Scholar] [CrossRef]
- Jacquelot, N.; Yamazaki, T.; Roberti, M.P.; Duong, C.P.M.; Andrews, M.C.; Verlingue, L.; Ferrere, G.; Becharef, S.; Vetizou, M.; Daillere, R.; et al. Sustained Type I interferon signaling as a mechanism of resistance to PD-1 blockade. Cell Res. 2019, 29, 846–861. [Google Scholar] [CrossRef]
- Hubert, M.; Gobbini, E.; Couillault, C.; Manh, T.V.; Doffin, A.C.; Berthet, J.; Rodriguez, C.; Ollion, V.; Kielbassa, J.; Sajous, C.; et al. IFN-III is selectively produced by cDC1 and predicts good clinical outcome in breast cancer. Sci. Immunol. 2020, 5, eaav3942. [Google Scholar] [CrossRef] [Green Version]
- Guilliams, M.; Crozat, K.; Henri, S.; Tamoutounour, S.; Grenot, P.; Devilard, E.; de Bovis, B.; Alexopoulou, L.; Dalod, M.; Malissen, B. Skin-draining lymph nodes contain dermis-derived CD103(-) dendritic cells that constitutively produce retinoic acid and induce Foxp3(+) regulatory T cells. Blood 2010, 115, 1958–1968. [Google Scholar] [CrossRef] [Green Version]
- Vitali, C.; Mingozzi, F.; Broggi, A.; Barresi, S.; Zolezzi, F.; Bayry, J.; Raimondi, G.; Zanoni, I.; Granucci, F. Migratory, and not lymphoid-resident, dendritic cells maintain peripheral self-tolerance and prevent autoimmunity via induction of iTreg cells. Blood 2012, 120, 1237–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fallarino, F.; Grohmann, U.; Hwang, K.W.; Orabona, C.; Vacca, C.; Bianchi, R.; Belladonna, M.L.; Fioretti, M.C.; Alegre, M.L.; Puccetti, P. Modulation of tryptophan catabolism by regulatory T cells. Nat. Immunol. 2003, 4, 1206–1212. [Google Scholar] [CrossRef] [PubMed]
- Orabona, C.; Pallotta, M.T.; Volpi, C.; Fallarino, F.; Vacca, C.; Bianchi, R.; Belladonna, M.L.; Fioretti, M.C.; Grohmann, U.; Puccetti, P. SOCS3 drives proteasomal degradation of indoleamine 2,3-dioxygenase (IDO) and antagonizes IDO-dependent tolerogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 20828–20833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orabona, C.; Tomasello, E.; Fallarino, F.; Bianchi, R.; Volpi, C.; Bellocchio, S.; Romani, L.; Fioretti, M.C.; Vivier, E.; Puccetti, P.; et al. Enhanced tryptophan catabolism in the absence of the molecular adapter DAP12. Eur. J. Immunol. 2005, 35, 3111–3118. [Google Scholar] [CrossRef]
- Belladonna, M.L.; Volpi, C.; Bianchi, R.; Vacca, C.; Orabona, C.; Pallotta, M.T.; Boon, L.; Gizzi, S.; Fioretti, M.C.; Grohmann, U.; et al. Cutting edge: Autocrine TGF-beta sustains default tolerogenesis by IDO-competent dendritic cells. J. Immunol. (Baltim. Md. 1950) 2008, 181, 5194–5198. [Google Scholar] [CrossRef] [Green Version]
- Banzola, I.; Mengus, C.; Wyler, S.; Hudolin, T.; Manzella, G.; Chiarugi, A.; Boldorini, R.; Sais, G.; Schmidli, T.S.; Chiffi, G.; et al. Expression of Indoleamine 2,3-Dioxygenase Induced by IFN-γ and TNF-α as Potential Biomarker of Prostate Cancer Progression. Front. Immunol. 2018, 9, 1051. [Google Scholar] [CrossRef] [Green Version]
- Fox, J.M.; Crabtree, J.M.; Sage, L.K.; Tompkins, S.M.; Tripp, R.A. Interferon Lambda Upregulates IDO1 Expression in Respiratory Epithelial Cells After Influenza Virus Infection. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2015, 35, 554–562. [Google Scholar] [CrossRef] [Green Version]
- Guillemin, G.J.; Kerr, S.J.; Pemberton, L.A.; Smith, D.G.; Smythe, G.A.; Armati, P.J.; Brew, B.J. IFN-beta1b induces kynurenine pathway metabolism in human macrophages: Potential implications for multiple sclerosis treatment. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2001, 21, 1097–1101. [Google Scholar] [CrossRef]
- Munn, D.H.; Mellor, A.L. IDO in the Tumor Microenvironment: Inflammation, Counter-Regulation, and Tolerance. Trends Immunol. 2016, 37, 193–207. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, J.C.; Andre, C.; Wang, Y.; Lawson, M.A.; Szegedi, S.S.; Lestage, J.; Castanon, N.; Kelley, K.W.; Dantzer, R. Interferon-gamma and tumor necrosis factor-alpha mediate the upregulation of indoleamine 2,3-dioxygenase and the induction of depressive-like behavior in mice in response to bacillus Calmette-Guerin. J. Neurosci. 2009, 29, 4200–4209. [Google Scholar] [CrossRef]
- Harden, J.L.; Egilmez, N.K. Indoleamine 2,3-dioxygenase and dendritic cell tolerogenicity. Immunol. Investig. 2012, 41, 738–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellor, A.L.; Munn, D.H. IDO expression by dendritic cells: Tolerance and tryptophan catabolism. Nat. Rev. Immunol. 2004, 4, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Munn, D.H.; Zhou, M.; Attwood, J.T.; Bondarev, I.; Conway, S.J.; Marshall, B.; Brown, C.; Mellor, A.L. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science (N. Y.) 1998, 281, 1191–1193. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Cao, X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell 2016, 30, 668–681. [Google Scholar] [CrossRef] [Green Version]
- Roberti, M.P.; Yonekura, S.; Duong, C.P.M.; Picard, M.; Ferrere, G.; Tidjani Alou, M.; Rauber, C.; Iebba, V.; Lehmann, C.H.K.; Amon, L.; et al. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nat. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Salmon, H.; Remark, R.; Gnjatic, S.; Merad, M. Host tissue determinants of tumour immunity. Nat. Rev. Cancer 2019, 19, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Gajewski, T.F. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat. Rev. Cancer 2018, 18, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Broz, M.L.; Binnewies, M.; Boldajipour, B.; Nelson, A.E.; Pollack, J.L.; Erle, D.J.; Barczak, A.; Rosenblum, M.D.; Daud, A.; Barber, D.L.; et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell 2014, 26, 638–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, S.; Efremova, M.; Riedel, A.; Mahata, B.; Pramanik, J.; Huuhtanen, J.; Kar, G.; Vento-Tormo, R.; Hagai, T.; Chen, X.; et al. Single-Cell RNA Sequencing Reveals a Dynamic Stromal Niche That Supports Tumor Growth. Cell Rep. 2020, 31, 107628. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.W.; Broz, M.L.; Binnewies, M.; Headley, M.B.; Nelson, A.E.; Wolf, D.M.; Kaisho, T.; Bogunovic, D.; Bhardwaj, N.; Krummel, M.F. Critical Role for CD103(+)/CD141(+) Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell 2016, 30, 324–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmon, H.; Idoyaga, J.; Rahman, A.; Leboeuf, M.; Remark, R.; Jordan, S.; Casanova-Acebes, M.; Khudoynazarova, M.; Agudo, J.; Tung, N.; et al. Expansion and Activation of CD103(+) Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity 2016, 44, 924–938. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Paulete, A.R.; Cueto, F.J.; Martinez-Lopez, M.; Labiano, S.; Morales-Kastresana, A.; Rodriguez-Ruiz, M.E.; Jure-Kunkel, M.; Azpilikueta, A.; Aznar, M.A.; Quetglas, J.I.; et al. Cancer Immunotherapy with Immunomodulatory Anti-CD137 and Anti-PD-1 Monoclonal Antibodies Requires BATF3-Dependent Dendritic Cells. Cancer Discov. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Spranger, S.; Dai, D.; Horton, B.; Gajewski, T.F. Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell 2017, 31, 711–723 e714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruffell, B.; Chang-Strachan, D.; Chan, V.; Rosenbusch, A.; Ho, C.M.; Pryer, N.; Daniel, D.; Hwang, E.S.; Rugo, H.S.; Coussens, L.M. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell 2014, 26, 623–637. [Google Scholar] [CrossRef] [Green Version]
- Binnewies, M.; Mujal, A.M.; Pollack, J.L.; Combes, A.J.; Hardison, E.A.; Barry, K.C.; Tsui, J.; Ruhland, M.K.; Kersten, K.; Abushawish, M.A.; et al. Unleashing Type-2 Dendritic Cells to Drive Protective Antitumor CD4(+) T Cell Immunity. Cell 2019, 177, 556–571.e516. [Google Scholar] [CrossRef]
- Headley, M.B.; Bins, A.; Nip, A.; Roberts, E.W.; Looney, M.R.; Gerard, A.; Krummel, M.F. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature 2016, 531, 513–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barry, K.C.; Hsu, J.; Broz, M.L.; Cueto, F.J.; Binnewies, M.; Combes, A.J.; Nelson, A.E.; Loo, K.; Kumar, R.; Rosenblum, M.D.; et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat. Med. 2018, 24, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.A.; Fearon, D.T. T cell exclusion, immune privilege, and the tumor microenvironment. Science (N. Y.) 2015, 348, 74–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villablanca, E.J.; Raccosta, L.; Zhou, D.; Fontana, R.; Maggioni, D.; Negro, A.; Sanvito, F.; Ponzoni, M.; Valentinis, B.; Bregni, M.; et al. Tumor-mediated liver X receptor-alpha activation inhibits CC chemokine receptor-7 expression on dendritic cells and dampens antitumor responses. Nat. Med. 2010, 16, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Bao, R.; Gajewski, T.F. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015, 523, 231–235. [Google Scholar] [CrossRef]
- Bol, K.F.; Schreibelt, G.; Rabold, K.; Wculek, S.K.; Schwarze, J.K.; Dzionek, A.; Teijeira, A.; Kandalaft, L.E.; Romero, P.; Coukos, G.; et al. The clinical application of cancer immunotherapy based on naturally circulating dendritic cells. J. Immunother. Cancer 2019, 7, 109. [Google Scholar] [CrossRef]
- Le Gall, C.M.; Weiden, J.; Eggermont, L.J.; Figdor, C.G. Dendritic cells in cancer immunotherapy. Nat. Mater. 2018, 17, 474–475. [Google Scholar] [CrossRef]
- Steinman, R.M.; Cohn, Z.A. Identification of a novel cell type in peripheral lymphoid organs of mice. II. Functional properties in vitro. J. Exp. Med. 1974, 139, 380–397. [Google Scholar] [CrossRef] [Green Version]
- Wimmers, F.; Schreibelt, G.; Skold, A.E.; Figdor, C.G.; De Vries, I.J. Paradigm Shift in Dendritic Cell-Based Immunotherapy: From in vitro Generated Monocyte-Derived DCs to Naturally Circulating DC Subsets. Front. Immunol. 2014, 5, 165. [Google Scholar] [CrossRef] [PubMed]
- Dorrie, J.; Schaft, N.; Schuler, G.; Schuler-Thurner, B. Therapeutic Cancer Vaccination with Ex vivo RNA-Transfected Dendritic Cells-An Update. Pharmaceutics 2020, 12, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sallusto, F.; Lanzavecchia, A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 1994, 179, 1109–1118. [Google Scholar] [CrossRef] [Green Version]
- Schuler-Thurner, B.; Schultz, E.S.; Berger, T.G.; Weinlich, G.; Ebner, S.; Woerl, P.; Bender, A.; Feuerstein, B.; Fritsch, P.O.; Romani, N.; et al. Rapid induction of tumor-specific type 1 T helper cells in metastatic melanoma patients by vaccination with mature, cryopreserved, peptide-loaded monocyte-derived dendritic cells. J. Exp. Med. 2002, 195, 1279–1288. [Google Scholar] [CrossRef]
- de Vries, I.J.M.; Lesterhuis, W.J.; Scharenborg, N.M.; Engelen, L.P.H.; Ruiter, D.J.; Gerritsen, M.-J.P.; Croockewit, S.; Britten, C.M.; Torensma, R.; Adema, G.J.; et al. Maturation of Dendritic Cells Is a Prerequisite for Inducing Immune Responses in Advanced Melanoma Patients. Clin. Cancer Res. 2003, 9, 5091. [Google Scholar]
- Dhodapkar, M.V.; Steinman, R.M.; Krasovsky, J.; Munz, C.; Bhardwaj, N. Antigen-specific inhibition of effector T cell function in humans after injection of immature dendritic cells. J. Exp. Med. 2001, 193, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Dhodapkar, M.V.; Steinman, R.M.; Sapp, M.; Desai, H.; Fossella, C.; Krasovsky, J.; Donahoe, S.M.; Dunbar, P.R.; Cerundolo, V.; Nixon, D.F.; et al. Rapid generation of broad T-cell immunity in humans after a single injection of mature dendritic cells. J. Clin. Investig. 1999, 104, 173–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonuleit, H.; Kuhn, U.; Muller, G.; Steinbrink, K.; Paragnik, L.; Schmitt, E.; Knop, J.; Enk, A.H. Pro-inflammatory cytokines and prostaglandins induce maturation of potent immunostimulatory dendritic cells under fetal calf serum-free conditions. Eur. J. Immunol. 1997, 27, 3135–3142. [Google Scholar] [CrossRef]
- Romani, N.; Reider, D.; Heuer, M.; Ebner, S.; Kampgen, E.; Eibl, B.; Niederwieser, D.; Schuler, G. Generation of mature dendritic cells from human blood. An improved method with special regard to clinical applicability. J. Immunol. Methods 1996, 196, 137–151. [Google Scholar] [CrossRef]
- Chang, A.E.; Redman, B.G.; Whitfield, J.R.; Nickoloff, B.J.; Braun, T.M.; Lee, P.P.; Geiger, J.D.; Mule, J.J. A phase I trial of tumor lysate-pulsed dendritic cells in the treatment of advanced cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2002, 8, 1021–1032. [Google Scholar]
- Condon, C.; Watkins, S.C.; Celluzzi, C.M.; Thompson, K.; Falo, L.D., Jr. DNA-based immunization by in vivo transfection of dendritic cells. Nat. Med. 1996, 2, 1122–1128. [Google Scholar] [CrossRef]
- Nestle, F.O.; Alijagic, S.; Gilliet, M.; Sun, Y.; Grabbe, S.; Dummer, R.; Burg, G.; Schadendorf, D. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nat. Med. 1998, 4, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Schaft, N.; Wellner, V.; Wohn, C.; Schuler, G.; Dorrie, J. CD8(+) T-cell priming and boosting: More antigen-presenting DC, or more antigen per DC? Cancer Immunol. Immunother. 2013, 62, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Schuler, G. Dendritic cells in cancer immunotherapy. Eur. J. Immunol. 2010, 40, 2123–2130. [Google Scholar] [CrossRef] [PubMed]
- Schuler, P.J.; Harasymczuk, M.; Visus, C.; Deleo, A.; Trivedi, S.; Lei, Y.; Argiris, A.; Gooding, W.; Butterfield, L.H.; Whiteside, T.L.; et al. Phase I dendritic cell p53 peptide vaccine for head and neck cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 2433–2444. [Google Scholar] [CrossRef] [Green Version]
- Tada, F.; Abe, M.; Hirooka, M.; Ikeda, Y.; Hiasa, Y.; Lee, Y.; Jung, N.C.; Lee, W.B.; Lee, H.S.; Bae, Y.S.; et al. Phase I/II study of immunotherapy using tumor antigen-pulsed dendritic cells in patients with hepatocellular carcinoma. Int. J. Oncol. 2012, 41, 1601–1609. [Google Scholar] [CrossRef] [Green Version]
- Osada, T.; Nagaoka, K.; Takahara, M.; Yang, X.Y.; Liu, C.X.; Guo, H.; Roy Choudhury, K.; Hobeika, A.; Hartman, Z.; Morse, M.A.; et al. Precision cancer immunotherapy: Optimizing dendritic cell-based strategies to induce tumor antigen-specific T-cell responses against individual patient tumors. J. Immunother. (Hagerstown Md. 1997) 2015, 38, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Vik-Mo, E.O.; Nyakas, M.; Mikkelsen, B.V.; Moe, M.C.; Due-Tonnesen, P.; Suso, E.M.; Saeboe-Larssen, S.; Sandberg, C.; Brinchmann, J.E.; Helseth, E.; et al. Therapeutic vaccination against autologous cancer stem cells with mRNA-transfected dendritic cells in patients with glioblastoma. Cancer Immunol. Immunother. 2013, 62, 1499–1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosch, N.C.; Voll, R.E.; Voskens, C.J.; Gross, S.; Seliger, B.; Schuler, G.; Schaft, N.; Dorrie, J. NF-kappaB activation triggers NK-cell stimulation by monocyte-derived dendritic cells. Ther. Adv. Med. Oncol. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Gerer, K.F.; Erdmann, M.; Hadrup, S.R.; Lyngaa, R.; Martin, L.M.; Voll, R.E.; Schuler-Thurner, B.; Schuler, G.; Schaft, N.; Hoyer, S.; et al. Preclinical evaluation of NF-kappaB-triggered dendritic cells expressing the viral oncogenic driver of Merkel cell carcinoma for therapeutic vaccination. Ther. Adv. Med. Oncol. 2017, 9, 451–464. [Google Scholar] [CrossRef] [Green Version]
- Pfeiffer, I.A.; Hoyer, S.; Gerer, K.F.; Voll, R.E.; Knippertz, I.; Guckel, E.; Schuler, G.; Schaft, N.; Dorrie, J. Triggering of NF-kappaB in cytokine-matured human DCs generates superior DCs for T-cell priming in cancer immunotherapy. Eur. J. Immunol. 2014, 44, 3413–3428. [Google Scholar] [CrossRef]
- Aerts, J.; de Goeje, P.L.; Cornelissen, R.; Kaijen-Lambers, M.E.H.; Bezemer, K.; van der Leest, C.H.; Mahaweni, N.M.; Kunert, A.; Eskens, F.; Waasdorp, C.; et al. Autologous Dendritic Cells Pulsed with Allogeneic Tumor Cell Lysate in Mesothelioma: From Mouse to Human. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 766–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fucikova, J.; Podrazil, M.; Jarolim, L.; Bilkova, P.; Hensler, M.; Becht, E.; Gasova, Z.; Klouckova, J.; Kayserova, J.; Horvath, R.; et al. Phase I/II trial of dendritic cell-based active cellular immunotherapy with DCVAC/PCa in patients with rising PSA after primary prostatectomy or salvage radiotherapy for the treatment of prostate cancer. Cancer Immunol. Immunother. 2018, 67, 89–100. [Google Scholar] [CrossRef]
- Ge, C.; Li, R.; Song, H.; Geng, T.; Yang, J.; Tan, Q.; Song, L.; Wang, Y.; Xue, Y.; Li, Z.; et al. Phase I clinical trial of a novel autologous modified-DC vaccine in patients with resected NSCLC. BMC Cancer 2017, 17, 884. [Google Scholar] [CrossRef]
- Lee, J.H.; Tak, W.Y.; Lee, Y.; Heo, M.K.; Song, J.S.; Kim, H.Y.; Park, S.Y.; Bae, S.H.; Lee, J.H.; Heo, J.; et al. Adjuvant immunotherapy with autologous dendritic cells for hepatocellular carcinoma, randomized phase II study. Oncoimmunology 2017, 6, e1328335. [Google Scholar] [CrossRef] [Green Version]
- Anguille, S.; Van de Velde, A.L.; Smits, E.L.; Van Tendeloo, V.F.; Juliusson, G.; Cools, N.; Nijs, G.; Stein, B.; Lion, E.; Van Driessche, A.; et al. Dendritic cell vaccination as postremission treatment to prevent or delay relapse in acute myeloid leukemia. Blood 2017, 130, 1713–1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gross, S.; Erdmann, M.; Haendle, I.; Voland, S.; Berger, T.; Schultz, E.; Strasser, E.; Dankerl, P.; Janka, R.; Schliep, S.; et al. Twelve-year survival and immune correlates in dendritic cell-vaccinated melanoma patients. JCI Insight. 2017, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowenfeld, L.; Mick, R.; Datta, J.; Xu, S.; Fitzpatrick, E.; Fisher, C.S.; Fox, K.R.; DeMichele, A.; Zhang, P.J.; Weinstein, S.P.; et al. Dendritic Cell Vaccination Enhances Immune Responses and Induces Regression of HER2(pos) DCIS Independent of Route: Results of Randomized Selection Design Trial. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 2961–2971. [Google Scholar] [CrossRef] [Green Version]
- Rains, N.; Cannan, R.J.; Chen, W.; Stubbs, R.S. Development of a dendritic cell (DC)-based vaccine for patients with advanced colorectal cancer. Hepatogastroenterology 2001, 48, 347–351. [Google Scholar]
- Nair, S.K.; Morse, M.; Boczkowski, D.; Cumming, R.I.; Vasovic, L.; Gilboa, E.; Lyerly, H.K. Induction of tumor-specific cytotoxic T lymphocytes in cancer patients by autologous tumor RNA-transfected dendritic cells. Ann. Surg. 2002, 235, 540–549. [Google Scholar] [CrossRef]
- Dannull, J.; Su, Z.; Rizzieri, D.; Yang, B.K.; Coleman, D.; Yancey, D.; Zhang, A.; Dahm, P.; Chao, N.; Gilboa, E.; et al. Enhancement of vaccine-mediated antitumor immunity in cancer patients after depletion of regulatory T cells. J. Clin. Investig. 2005, 115, 3623–3633. [Google Scholar] [CrossRef]
- Caruso, D.A.; Orme, L.M.; Neale, A.M.; Radcliff, F.J.; Amor, G.M.; Maixner, W.; Downie, P.; Hassall, T.E.; Tang, M.L.; Ashley, D.M. Results of a phase 1 study utilizing monocyte-derived dendritic cells pulsed with tumor RNA in children and young adults with brain cancer. Neuro Oncol. 2004, 6, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Caruso, D.A.; Orme, L.M.; Amor, G.M.; Neale, A.M.; Radcliff, F.J.; Downie, P.; Tang, M.L.; Ashley, D.M. Results of a Phase I study utilizing monocyte-derived dendritic cells pulsed with tumor RNA in children with Stage 4 neuroblastoma. Cancer 2005, 103, 1280–1291. [Google Scholar] [CrossRef]
- Shindo, Y.; Hazama, S.; Maeda, Y.; Matsui, H.; Iida, M.; Suzuki, N.; Yoshimura, K.; Ueno, T.; Yoshino, S.; Sakai, K.; et al. Adoptive immunotherapy with MUC1-mRNA transfected dendritic cells and cytotoxic lymphocytes plus gemcitabine for unresectable pancreatic cancer. J. Transl. Med. 2014, 12, 175. [Google Scholar] [CrossRef] [Green Version]
- Hsu, F.J.; Benike, C.; Fagnoni, F.; Liles, T.M.; Czerwinski, D.; Taidi, B.; Engleman, E.G.; Levy, R. Vaccination of patients with B-cell lymphoma using autologous antigen-pulsed dendritic cells. Nat. Med. 1996, 2, 52–58. [Google Scholar] [CrossRef]
- Batich, K.A.; Reap, E.A.; Archer, G.E.; Sanchez-Perez, L.; Nair, S.K.; Schmittling, R.J.; Norberg, P.; Xie, W.; Herndon, J.E., 2nd; Healy, P.; et al. Long-term Survival in Glioblastoma with Cytomegalovirus pp65-Targeted Vaccination. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 1898–1909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palucka, A.K.; Ueno, H.; Connolly, J.; Kerneis-Norvell, F.; Blanck, J.P.; Johnston, D.A.; Fay, J.; Banchereau, J. Dendritic cells loaded with killed allogeneic melanoma cells can induce objective clinical responses and MART-1 specific CD8+ T-cell immunity. J. Immunother. (Hagerstown Md. 1997) 2006, 29, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Engell-Noerregaard, L.; Hansen, T.H.; Andersen, M.H.; Thor Straten, P.; Svane, I.M. Review of clinical studies on dendritic cell-based vaccination of patients with malignant melanoma: Assessment of correlation between clinical response and vaccine parameters. Cancer Immunol. Immunother. 2009, 58, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Granier, C.; De Guillebon, E.; Blanc, C.; Roussel, H.; Badoual, C.; Colin, E.; Saldmann, A.; Gey, A.; Oudard, S.; Tartour, E. Mechanisms of action and rationale for the use of checkpoint inhibitors in cancer. ESMO Open 2017, 2, e000213. [Google Scholar] [CrossRef] [Green Version]
- Stronen, E.; Toebes, M.; Kelderman, S.; van Buuren, M.M.; Yang, W.; van Rooij, N.; Donia, M.; Boschen, M.L.; Lund-Johansen, F.; Olweus, J.; et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science (N. Y.) 2016, 352, 1337–1341. [Google Scholar] [CrossRef]
- Wei, S.C.; Duffy, C.R.; Allison, J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018, 8, 1069–1086. [Google Scholar] [CrossRef] [Green Version]
- Menendez, P.; Prosper, F.; Bueno, C.; Arbona, C.; San Miguel, J.F.; Garcia-Conde, J.; Sola, C.; Hornedo, J.; Cortes-Funes, H.; Orfao, A. Sequential analysis of CD34+ and CD34- cell subsets in peripheral blood and leukapheresis products from breast cancer patients mobilized with SCF plus G-CSF and cyclophosphamide. Leukemia 2001, 15, 430–439. [Google Scholar] [CrossRef] [Green Version]
- Paczesny, S.; Li, Y.P.; Li, N.; Latger-Cannard, V.; Marchal, L.; Ou-Yang, J.P.; Bordigoni, P.; Stoltz, J.F.; Eljaafari, A. Efficient generation of CD34+ progenitor-derived dendritic cells from G-CSF-mobilized peripheral mononuclear cells does not require hematopoietic stem cell enrichment. J. Leukoc. Boil. 2007, 81, 957–967. [Google Scholar] [CrossRef]
- Ratta, M.; Rondelli, D.; Fortuna, A.; Curti, A.; Fogli, M.; Fagnoni, F.; Martinelli, G.; Terragna, C.; Tura, S.; Lemoli, R.M. Generation and functional characterization of human dendritic cells derived from CD34 cells mobilized into peripheral blood: Comparison with bone marrow CD34+ cells. Br. J. Haematol. 1998, 101, 756–765. [Google Scholar] [CrossRef]
- Caux, C.; Massacrier, C.; Vanbervliet, B.; Dubois, B.; Durand, I.; Cella, M.; Lanzavecchia, A.; Banchereau, J. CD34+ hematopoietic progenitors from human cord blood differentiate along two independent dendritic cell pathways in response to granulocyte-macrophage colony-stimulating factor plus tumor necrosis factor alpha: II. Functional analysis. Blood 1997, 90, 1458–1470. [Google Scholar] [CrossRef]
- Caux, C.; Vanbervliet, B.; Massacrier, C.; Dezutter-Dambuyant, C.; de Saint-Vis, B.; Jacquet, C.; Yoneda, K.; Imamura, S.; Schmitt, D.; Banchereau, J. CD34+ hematopoietic progenitors from human cord blood differentiate along two independent dendritic cell pathways in response to GM-CSF+TNF alpha. J. Exp. Med. 1996, 184, 695–706. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, J.; Palucka, A.K.; Dhodapkar, M.; Burkeholder, S.; Taquet, N.; Rolland, A.; Taquet, S.; Coquery, S.; Wittkowski, K.M.; Bhardwaj, N.; et al. Immune and clinical responses in patients with metastatic melanoma to CD34(+) progenitor-derived dendritic cell vaccine. Cancer Res. 2001, 61, 6451–6458. [Google Scholar] [PubMed]
- Palucka, A.K.; Ueno, H.; Fay, J.; Banchereau, J. Dendritic cells: A critical player in cancer therapy? J. Immunother. (Hagerstown Md. 1997) 2008, 31, 793–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palucka, K.; Banchereau, J. Cancer immunotherapy via dendritic cells. Nat. Rev. Cancer 2012, 12, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Ratzinger, G.; Baggers, J.; de Cos, M.A.; Yuan, J.D.; Dao, T.; Reagan, J.L.; Munz, C.; Heller, G.; Young, J.W. Mature human langerhans cells derived from CD34(+) hematopoietic progenitors stimulate greater cytolytic. T lymphocyte activity in the absence of bioactive IL-12p70, by either single peptide presentation or cross-priming, than do dermal-interstitial or monocyte-derived dendritic cells. J. Immunol. 2005, 174, 2780–2791. [Google Scholar] [CrossRef]
- Yuan, J.; Latouche, J.B.; Reagan, J.L.; Heller, G.; Riviere, I.; Sadelain, M.; Young, J.W. Langerhans cells derived from genetically modified human CD34+ hemopoietic progenitors are more potent than peptide-pulsed Langerhans cells for inducing antigen-specific CD8+ cytolytic T lymphocyte responses. J. Immunol. (Baltim. Md. 1950) 2005, 174, 758–766. [Google Scholar] [CrossRef] [Green Version]
- Fay, J.W.; Palucka, A.K.; Paczesny, S.; Dhodapkar, M.; Johnston, D.A.; Burkeholder, S.; Ueno, H.; Banchereau, J. Long-term outcomes in patients with metastatic melanoma vaccinated with melanoma peptide-pulsed CD34(+) progenitor-derived dendritic cells. Cancer Immunol. Immunother. 2006, 55, 1209–1218. [Google Scholar] [CrossRef]
- Paczesny, S.; Banchereau, J.; Wittkowski, K.M.; Saracino, G.; Fay, J.; Palucka, A.K. Expansion of melanoma-specific cytolytic CD8+ T cell precursors in patients with metastatic melanoma vaccinated with CD34+ progenitor-derived dendritic cells. J. Exp. Med. 2004, 199, 1503–1511. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Breton, G.; Aljoufi, A.; Zhou, Y.J.; Puhr, S.; Nussenzweig, M.C.; Liu, K. Clonal analysis of human dendritic cell progenitor using a stromal cell culture. J. Immunol. Methods 2015, 425, 21–26. [Google Scholar] [CrossRef] [Green Version]
- Anselmi, G.; Vaivode, K.; Dutertre, C.A.; Bourdely, P.; Missolo-Koussou, Y.; Newell, E.; Hickman, O.; Wood, K.; Saxena, A.; Helft, J.; et al. Engineered niches support the development of human dendritic cells in humanized mice. Nat. Commun. 2020, 11, 2054. [Google Scholar] [CrossRef]
- Breton, G.; Lee, J.; Zhou, Y.J.; Schreiber, J.J.; Keler, T.; Puhr, S.; Anandasabapathy, N.; Schlesinger, S.; Caskey, M.; Liu, K.; et al. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J. Exp. Med. 2015, 212, 401–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marroquin, C.E.; Westwood, J.A.; Lapointe, R.; Mixon, A.; Wunderlich, J.R.; Caron, D.; Rosenberg, S.A.; Hwu, P. Mobilization of dendritic cell precursors in patients with cancer by flt3 ligand allows the generation of higher yields of cultured dendritic cells. J. Immunother. (Hagerstown Md. 1997) 2002, 25, 278–288. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, N.; Pavlick, A.C.; Ernstoff, M.S.; Hanks, B.A.; Albertini, M.R.; Luke, J.J.; Yellin, M.J.; Keler, T.; Davis, T.A.; Crocker, A.; et al. A Phase II Randomized Study of CDX-1401, a Dendritic Cell Targeting NY-ESO-1 Vaccine, in Patients with Malignant Melanoma Pre-Treated with Recombinant CDX-301, a Recombinant Human Flt3 Ligand. J. Clin. Oncol. 2016, 34, 9589. [Google Scholar] [CrossRef]
- Schreibelt, G.; Bol, K.F.; Westdorp, H.; Wimmers, F.; Aarntzen, E.H.; Duiveman-de Boer, T.; van de Rakt, M.W.; Scharenborg, N.M.; de Boer, A.J.; Pots, J.M.; et al. Effective Clinical Responses in Metastatic Melanoma Patients after Vaccination with Primary Myeloid Dendritic Cells. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 2155–2166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prue, R.L.; Vari, F.; Radford, K.J.; Tong, H.; Hardy, M.Y.; D’Rozario, R.; Waterhouse, N.J.; Rossetti, T.; Coleman, R.; Tracey, C.; et al. A phase I clinical trial of CD1c (BDCA-1)+ dendritic cells pulsed with HLA-A*0201 peptides for immunotherapy of metastatic hormone refractory prostate cancer. J. Immunother. (Hagerstown Md. 1997) 2015, 38, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Sabado, R.L.; Balan, S.; Bhardwaj, N. Dendritic cell-based immunotherapy. Cell Res. 2017, 27, 74–95. [Google Scholar] [CrossRef] [Green Version]
- Huber, A.; Dammeijer, F.; Aerts, J.; Vroman, H. Current State of Dendritic Cell-Based Immunotherapy: Opportunities for in vitro Antigen Loading of Different DC Subsets? Front. Immunol. 2018, 9, 2804. [Google Scholar] [CrossRef]
- Kraal, G.; Breel, M.; Janse, M.; Bruin, G. Langerhans’ cells, veiled cells, and interdigitating cells in the mouse recognized by a monoclonal antibody. J. Exp. Med. 1986, 163, 981–997. [Google Scholar] [CrossRef] [Green Version]
- Nussenzweig, M.C.; Steinman, R.M.; Witmer, M.D.; Gutchinov, B. A monoclonal antibody specific for mouse dendritic cells. Proc. Natl. Acad. Sci. USA 1982, 79, 161–165. [Google Scholar] [CrossRef] [Green Version]
- Bonifaz, L.C.; Bonnyay, D.P.; Charalambous, A.; Darguste, D.I.; Fujii, S.; Soares, H.; Brimnes, M.K.; Moltedo, B.; Moran, T.M.; Steinman, R.M. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J. Exp. Med. 2004, 199, 815–824. [Google Scholar] [CrossRef] [Green Version]
- Cruz, L.J.; Rosalia, R.A.; Kleinovink, J.W.; Rueda, F.; Lowik, C.W.; Ossendorp, F. Targeting nanoparticles to CD40, DEC-205 or CD11c molecules on dendritic cells for efficient CD8(+) T cell response: A comparative study. J. Control. Release Off. J. Control. Release Soc. 2014, 192, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Price, J.D.; Hotta-Iwamura, C.; Zhao, Y.; Beauchamp, N.M.; Tarbell, K.V. DCIR2+ cDC2 DCs and Zbtb32 Restore CD4+ T-Cell Tolerance and Inhibit Diabetes. Diabetes 2015, 64, 3521–3531. [Google Scholar] [CrossRef] [Green Version]
- Tullett, K.M.; Leal Rojas, I.M.; Minoda, Y.; Tan, P.S.; Zhang, J.G.; Smith, C.; Khanna, R.; Shortman, K.; Caminschi, I.; Lahoud, M.H.; et al. Targeting CLEC9A delivers antigen to human CD141(+) DC for CD4(+) and CD8(+)T cell recognition. JCI Insight. 2016, 1, e87102. [Google Scholar] [CrossRef] [PubMed]
- Castro, F.V.; Tutt, A.L.; White, A.L.; Teeling, J.L.; James, S.; French, R.R.; Glennie, M.J. CD11c provides an effective immunotarget for the generation of both CD4 and CD8 T cell responses. Eur. J. Immunol. 2008, 38, 2263–2273. [Google Scholar] [CrossRef] [PubMed]
- Delneste, Y.; Magistrelli, G.; Gauchat, J.; Haeuw, J.; Aubry, J.; Nakamura, K.; Kawakami-Honda, N.; Goetsch, L.; Sawamura, T.; Bonnefoy, J.; et al. Involvement of LOX-1 in dendritic cell-mediated antigen cross-presentation. Immunity 2002, 17, 353–362. [Google Scholar] [CrossRef] [Green Version]
- Dickgreber, N.; Stoitzner, P.; Bai, Y.; Price, K.M.; Farrand, K.J.; Manning, K.; Angel, C.E.; Dunbar, P.R.; Ronchese, F.; Fraser, J.D.; et al. Targeting antigen to MHC class II molecules promotes efficient cross-presentation and enhances immunotherapy. J. Immunol. (Baltim. Md. 1950) 2009, 182, 1260–1269. [Google Scholar] [CrossRef] [PubMed]
- Macho-Fernandez, E.; Cruz, L.J.; Ghinnagow, R.; Fontaine, J.; Bialecki, E.; Frisch, B.; Trottein, F.; Faveeuw, C. Targeted delivery of alpha-galactosylceramide to CD8alpha+ dendritic cells optimizes type I NKT cell-based antitumor responses. J. Immunol. (Baltim. Md. 1950) 2014, 193, 961–969. [Google Scholar] [CrossRef] [Green Version]
- Mahnke, K.; Qian, Y.; Fondel, S.; Brueck, J.; Becker, C.; Enk, A.H. Targeting of antigens to activated dendritic cells in vivo cures metastatic melanoma in mice. Cancer Res. 2005, 65, 7007–7012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sancho, D.; Mourao-Sa, D.; Joffre, O.P.; Schulz, O.; Rogers, N.C.; Pennington, D.J.; Carlyle, J.R.; Reis e Sousa, C. Tumor therapy in mice via antigen targeting to a novel, DC-restricted C-type lectin. J. Clin. Investig. 2008, 118, 2098–2110. [Google Scholar] [CrossRef]
- Tagliani, E.; Guermonprez, P.; Sepulveda, J.; Lopez-Bravo, M.; Ardavin, C.; Amigorena, S.; Benvenuti, F.; Burrone, O.R. Selection of an antibody library identifies a pathway to induce immunity by targeting CD36 on steady-state CD8 alpha+ dendritic cells. J. Immunol. (Baltim. Md. 1950) 2008, 180, 3201–3209. [Google Scholar] [CrossRef] [Green Version]
- van Broekhoven, C.L.; Parish, C.R.; Demangel, C.; Britton, W.J.; Altin, J.G. Targeting dendritic cells with antigen-containing liposomes: A highly effective procedure for induction of antitumor immunity and for tumor immunotherapy. Cancer Res. 2004, 64, 4357–4365. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Wang, S.; Zhang, D.; Hou, S.; Qian, W.; Li, B.; Guo, H.; Kou, G.; He, J.; Wang, H.; et al. Targeted delivery of tumor antigens to activated dendritic cells via CD11c molecules induces potent antitumor immunity in mice. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 4612–4621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishna, M.; Nadler, S.G. Immunogenicity to Biotherapeutics—The Role of Anti-drug Immune Complexes. Front. Immunol. 2016, 7, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pratt, K.P. Anti-Drug Antibodies: Emerging Approaches to Predict, Reduce or Reverse Biotherapeutic Immunogenicity. Antibodies 2018, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowers, P.M.; Neben, T.Y.; Tomlinson, G.L.; Dalton, J.L.; Altobell, L.; Zhang, X.; Macomber, J.L.; Wu, B.F.; Toobian, R.M.; McConnell, A.D.; et al. Humanization of antibodies using heavy chain complementarity-determining region 3 grafting coupled with in vitro somatic hypermutation. J. Boil. Chem. 2013, 288, 7688–7696. [Google Scholar] [CrossRef] [Green Version]
- Clavero-Alvarez, A.; Di Mambro, T.; Perez-Gaviro, S.; Magnani, M.; Bruscolini, P. Humanization of Antibodies using a Statistical Inference Approach. Sci. Rep. 2018, 8, 14820. [Google Scholar] [CrossRef]
- Safdari, Y.; Farajnia, S.; Asgharzadeh, M.; Khalili, M. Antibody humanization methods—A review and update. Biotechnol. Genet. Eng. Rev. 2013, 29, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.C.; Liang, Q.; Ali, H.; Bayliss, L.; Beasley, A.; Bloomfield-Gerdes, T.; Bonoli, L.; Brown, R.; Campbell, J.; Carpenter, A.; et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat. Biotechnol. 2014, 32, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, L.E.; Karow, M.; Stevens, S.; Auerbach, W.; Poueymirou, W.T.; Yasenchak, J.; Frendewey, D.; Valenzuela, D.M.; Giallourakis, C.C.; Alt, F.W.; et al. Precise and in situ genetic humanization of 6 Mb of mouse immunoglobulin genes. Proc. Natl. Acad. Sci. USA 2014, 111, 5147–5152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendez, M.J.; Green, L.L.; Corvalan, J.R.; Jia, X.C.; Maynard-Currie, C.E.; Yang, X.D.; Gallo, M.L.; Louie, D.M.; Lee, D.V.; Erickson, K.L.; et al. Functional transplant of megabase human immunoglobulin loci recapitulates human antibody response in mice. Nat. Genet. 1997, 15, 146–156. [Google Scholar] [CrossRef]
- Murphy, A.J.; Macdonald, L.E.; Stevens, S.; Karow, M.; Dore, A.T.; Pobursky, K.; Huang, T.T.; Poueymirou, W.T.; Esau, L.; Meola, M.; et al. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc. Natl. Acad. Sci. USA 2014, 111, 5153–5158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruhns, P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood 2012, 119, 5640–5649. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, F.; Bruhns, P.; Horiuchi, K.; Ravetch, J.V. FcgammaRIV: A novel FcR with distinct IgG subclass specificity. Immunity 2005, 23, 41–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.F.; Sazinsky, S.L.; Houde, D.; DiLillo, D.J.; Bird, J.; Li, K.K.; Cheng, G.T.; Qiu, H.; Engen, J.R.; Ravetch, J.V.; et al. Engineering Aglycosylated IgG Variants with Wild-Type or Improved Binding Affinity to Human Fc Gamma RIIA and Fc Gamma RIIIAs. J. Mol. Biol. 2017, 429, 2528–2541. [Google Scholar] [CrossRef]
- Kao, D.; Danzer, H.; Collin, M.; Gross, A.; Eichler, J.; Stambuk, J.; Lauc, G.; Lux, A.; Nimmerjahn, F. A Monosaccharide Residue Is Sufficient to Maintain Mouse and Human IgG Subclass Activity and Directs IgG Effector Functions to Cellular Fc Receptors. Cell Rep. 2015, 13, 2376–2385. [Google Scholar] [CrossRef] [Green Version]
- Subedi, G.P.; Barb, A.W. The Structural Role of Antibody N-Glycosylation in Receptor Interactions. Structure 2015, 23, 1573–1583. [Google Scholar] [CrossRef] [Green Version]
- Walker, M.R.; Lund, J.; Thompson, K.M.; Jefferis, R. Aglycosylation of human IgG1 and IgG3 monoclonal antibodies can eliminate recognition by human cells expressing Fc gamma RI and/or Fc gamma RII receptors. Biochem. J. 1989, 259, 347–353. [Google Scholar] [CrossRef]
- Benito-Villalvilla, C.; Soria, I.; Subiza, J.L.; Palomares, O. Novel vaccines targeting dendritic cells by coupling allergoids to mannan. Allergo J. Int. 2018, 27, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Frenz, T.; Grabski, E.; Duran, V.; Hozsa, C.; Stepczynska, A.; Furch, M.; Gieseler, R.K.; Kalinke, U. Antigen presenting cell-selective drug delivery by glycan-decorated nanocarriers. Eur. J. pharm. Biopharm. Off. J. Arb. Fur Pharm. Verfahrenstechnik e.V 2015, 95, 13–17. [Google Scholar] [CrossRef]
- Johannssen, T.; Lepenies, B. Glycan-Based Cell Targeting To Modulate Immune Responses. Trends Biotechnol. 2017, 35, 334–346. [Google Scholar] [CrossRef] [Green Version]
- Wamhoff, E.C.; Schulze, J.; Bellmann, L.; Rentzsch, M.; Bachem, G.; Fuchsberger, F.F.; Rademacher, J.; Hermann, M.; Del Frari, B.; van Dalen, R.; et al. A Specific, Glycomimetic Langerin Ligand for Human Langerhans Cell Targeting. ACS Cent. Sci. 2019, 5, 808–820. [Google Scholar] [CrossRef] [Green Version]
- Donadei, A.; Gallorini, S.; Berti, F.; O’Hagan, D.T.; Adamo, R.; Baudner, B.C. Rational Design of Adjuvant for Skin Delivery: Conjugation of Synthetic beta-Glucan Dectin-1 Agonist to Protein Antigen. Mol. Pharm. 2015, 12, 1662–1672. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vallejo, J.J.; Bloem, K.; Knippels, L.M.; Garssen, J.; van Vliet, S.J.; van Kooyk, Y. The Consequences of Multiple Simultaneous C-Type Lectin-Ligand Interactions: DCIR Alters the Endo-Lysosomal Routing of DC-SIGN. Front. Immunol. 2015, 6, 87. [Google Scholar] [CrossRef] [Green Version]
- Streng-Ouwehand, I.; Ho, N.I.; Litjens, M.; Kalay, H.; Boks, M.A.; Cornelissen, L.A.; Kaur Singh, S.; Saeland, E.; Garcia-Vallejo, J.J.; Ossendorp, F.A.; et al. Glycan modification of antigen alters its intracellular routing in dendritic cells, promoting priming of T cells. eLife 2016, 5, e11765. [Google Scholar] [CrossRef] [Green Version]
- van Kooyk, Y.; Unger, W.W.; Fehres, C.M.; Kalay, H.; Garcia-Vallejo, J.J. Glycan-based DC-SIGN targeting vaccines to enhance antigen cross-presentation. Mol. Immunol. 2013, 55, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Wolfert, M.A.; Boons, G.J. Adaptive immune activation: Glycosylation does matter. Nat. Chem. Biol. 2013, 9, 776–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Platt, C.D.; Ma, J.K.; Chalouni, C.; Ebersold, M.; Bou-Reslan, H.; Carano, R.A.; Mellman, I.; Delamarre, L. Mature dendritic cells use endocytic receptors to capture and present antigens. Proc. Natl. Acad. Sci. USA 2010, 107, 4287–4292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourquin, C.; Anz, D.; Zwiorek, K.; Lanz, A.L.; Fuchs, S.; Weigel, S.; Wurzenberger, C.; von der Borch, P.; Golic, M.; Moder, S.; et al. Targeting CpG oligonucleotides to the lymph node by nanoparticles elicits efficient antitumoral immunity. J. Immunol. (Baltim. Md. 1950) 2008, 181, 2990–2998. [Google Scholar] [CrossRef] [PubMed]
- Kawagoe, T.; Takeuchi, O.; Takabatake, Y.; Kato, H.; Isaka, Y.; Tsujimura, T.; Akira, S. TANK is a negative regulator of Toll-like receptor signaling and is critical for the prevention of autoimmune nephritis. Nat. Immunol. 2009, 10, 965–972. [Google Scholar] [CrossRef] [Green Version]
- Storni, T.; Ruedl, C.; Schwarz, K.; Schwendener, R.A.; Renner, W.A.; Bachmann, M.F. Nonmethylated CG motifs packaged into virus-like particles induce protective cytotoxic T cell responses in the absence of systemic side effects. J. Immunol. (Baltim. Md. 1950) 2004, 172, 1777–1785. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Zhao, J.; Unkeless, J.C.; Feng, Z.H.; Xiong, H. TLR signaling by tumor and immune cells: A double-edged sword. Oncogene 2008, 27, 218–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blander, J.M.; Medzhitov, R. Toll-dependent selection of microbial antigens for presentation by dendritic cells. Nature 2006, 440, 808–812. [Google Scholar] [CrossRef]
- Lang, K.S.; Recher, M.; Junt, T.; Navarini, A.A.; Harris, N.L.; Freigang, S.; Odermatt, B.; Conrad, C.; Ittner, L.M.; Bauer, S.; et al. Toll-like receptor engagement converts T-cell autoreactivity into overt autoimmune disease. Nat. Med. 2005, 11, 138–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, K.H. TLR-dependent T cell activation in autoimmunity. Nat. Rev. Immunol. 2011, 11, 807–822. [Google Scholar] [CrossRef] [PubMed]
- Sacher, T.; Knolle, P.; Nichterlein, T.; Arnold, B.; Hammerling, G.J.; Limmer, A. CpG-ODN-induced inflammation is sufficient to cause T-cell-mediated autoaggression against hepatocytes. Eur. J. Immunol. 2002, 32, 3628–3637. [Google Scholar] [CrossRef]
- Barbuto, S.; Idoyaga, J.; Vila-Perello, M.; Longhi, M.P.; Breton, G.; Steinman, R.M.; Muir, T.W. Induction of innate and adaptive immunity by delivery of poly dA:dT to dendritic cells. Nat. Chem. Biol. 2013, 9, 250–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heit, A.; Schmitz, F.; O’Keeffe, M.; Staib, C.; Busch, D.H.; Wagner, H.; Huster, K.M. Protective CD8 T cell immunity triggered by CpG-protein conjugates competes with the efficacy of live vaccines. J. Immunol. (Baltim. Md. 1950) 2005, 174, 4373–4380. [Google Scholar] [CrossRef] [Green Version]
- Kreutz, M.; Giquel, B.; Hu, Q.; Abuknesha, R.; Uematsu, S.; Akira, S.; Nestle, F.O.; Diebold, S.S. Antibody-antigen-adjuvant conjugates enable co-delivery of antigen and adjuvant to dendritic cells in cis but only have partial targeting specificity. PLoS ONE 2012, 7, e40208. [Google Scholar] [CrossRef] [Green Version]
- Badiee, A.; Davies, N.; McDonald, K.; Radford, K.; Michiue, H.; Hart, D.; Kato, M. Enhanced delivery of immunoliposomes to human dendritic cells by targeting the multilectin receptor DEC-205. Vaccine 2007, 25, 4757–4766. [Google Scholar] [CrossRef]
- Brandao, J.G.; Scheper, R.J.; Lougheed, S.M.; Curiel, D.T.; Tillman, B.W.; Gerritsen, W.R.; van den Eertwegh, A.J.; Pinedo, H.M.; Haisma, H.J.; de Gruijl, T.D. CD40-targeted adenoviral gene transfer to dendritic cells through the use of a novel bispecific single-chain Fv antibody enhances cytotoxic T cell activation. Vaccine 2003, 21, 2268–2272. [Google Scholar] [CrossRef]
- Kwon, Y.J.; James, E.; Shastri, N.; Frechet, J.M. In vivo targeting of dendritic cells for activation of cellular immunity using vaccine carriers based on pH-responsive microparticles. Proc. Natl. Acad. Sci. USA 2005, 102, 18264–18268. [Google Scholar] [CrossRef] [Green Version]
- Shi, G.N.; Zhang, C.N.; Xu, R.; Niu, J.F.; Song, H.J.; Zhang, X.Y.; Wang, W.W.; Wang, Y.M.; Li, C.; Wei, X.Q.; et al. Enhanced antitumor immunity by targeting dendritic cells with tumor cell lysate-loaded chitosan nanoparticles vaccine. Biomaterials 2017, 113, 191–202. [Google Scholar] [CrossRef]
- Tacken, P.J.; Zeelenberg, I.S.; Cruz, L.J.; van Hout-Kuijer, M.A.; van de Glind, G.; Fokkink, R.G.; Lambeck, A.J.; Figdor, C.G. Targeted delivery of TLR ligands to human and mouse dendritic cells strongly enhances adjuvanticity. Blood 2011, 118, 6836–6844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomann, J.S.; Heurtault, B.; Weidner, S.; Braye, M.; Beyrath, J.; Fournel, S.; Schuber, F.; Frisch, B. Antitumor activity of liposomal ErbB2/HER2 epitope peptide-based vaccine constructs incorporating TLR agonists and mannose receptor targeting. Biomaterials 2011, 32, 4574–4583. [Google Scholar] [CrossRef] [PubMed]
- Mukai, Y.; Yoshinaga, T.; Yoshikawa, M.; Matsuo, K.; Yoshikawa, T.; Matsuo, K.; Niki, K.; Yoshioka, Y.; Okada, N.; Nakagawa, S. Induction of endoplasmic reticulum-endosome fusion for antigen cross-presentation induced by poly (gamma-glutamic acid) nanoparticles. J. Immunol. (Baltim. Md. 1950) 2011, 187, 6249–6255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, T.; Moriguchi, R.; Kogure, K.; Shastri, N.; Harashima, H. Efficient MHC class I presentation by controlled intracellular trafficking of antigens in octaarginine-modified liposomes. Mol. Ther. J. Am. Soc. Gene Ther. 2008, 16, 1507–1514. [Google Scholar] [CrossRef]
- Yuba, E.; Harada, A.; Sakanishi, Y.; Watarai, S.; Kono, K. A liposome-based antigen delivery system using pH-sensitive fusogenic polymers for cancer immunotherapy. Biomaterials 2013, 34, 3042–3052. [Google Scholar] [CrossRef]
- Wang, H.; Yu, X.; Guo, C.; Zuo, D.; Fisher, P.B.; Subjeck, J.R.; Wang, X.Y. Enhanced endoplasmic reticulum entry of tumor antigen is crucial for cross-presentation induced by dendritic cell-targeted vaccination. J. Immunol. (Baltim. Md. 1950) 2013, 191, 6010–6021. [Google Scholar] [CrossRef] [Green Version]
- Basha, G.; Novobrantseva, T.I.; Rosin, N.; Tam, Y.Y.; Hafez, I.M.; Wong, M.K.; Sugo, T.; Ruda, V.M.; Qin, J.; Klebanov, B.; et al. Influence of cationic lipid composition on gene silencing properties of lipid nanoparticle formulations of siRNA in antigen-presenting cells. Mol. Ther. J. Am. Soc. Gene Ther. 2011, 19, 2186–2200. [Google Scholar] [CrossRef]
- Kenworthy, R.; Lambert, D.; Yang, F.; Wang, N.; Chen, Z.; Zhu, H.; Zhu, F.; Liu, C.; Li, K.; Tang, H. Short-hairpin RNAs delivered by lentiviral vector transduction trigger RIG-I-mediated IFN activation. Nucleic Acids Res. 2009, 37, 6587–6599. [Google Scholar] [CrossRef]
- Kreutz, M.; Tacken, P.J.; Figdor, C.G. Targeting dendritic cells--why bother? Blood 2013, 121, 2836–2844. [Google Scholar] [CrossRef] [PubMed]
- McCullough, K.C.; Bassi, I.; Demoulins, T.; Thomann-Harwood, L.J.; Ruggli, N. Functional RNA delivery targeted to dendritic cells by synthetic nanoparticles. Ther. Deliv. 2012, 3, 1077–1099. [Google Scholar] [CrossRef] [Green Version]
- Paulis, L.E.; Mandal, S.; Kreutz, M.; Figdor, C.G. Dendritic cell-based nanovaccines for cancer immunotherapy. Curr. Opin. Immunol. 2013, 25, 389–395. [Google Scholar] [CrossRef]
- van Dinther, D.; Stolk, D.A.; van de Ven, R.; van Kooyk, Y.; de Gruijl, T.D.; den Haan, J.M.M. Targeting C-type lectin receptors: A high-carbohydrate diet for dendritic cells to improve cancer vaccines. J. Leukoc. Boil. 2017, 102, 1017–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, M.L.; Schnorfeil, F.M.; Brocker, T. MicroRNAs regulate dendritic cell differentiation and function. J. Immunol. (Baltim. Md. 1950) 2011, 187, 3911–3917. [Google Scholar] [CrossRef]
- Zhou, H.; Wu, L. The development and function of dendritic cell populations and their regulation by miRNAs. Protein Cell 2017, 8, 501–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brigl, M.; Tatituri, R.V.; Watts, G.F.; Bhowruth, V.; Leadbetter, E.A.; Barton, N.; Cohen, N.R.; Hsu, F.F.; Besra, G.S.; Brenner, M.B. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J. Exp. Med. 2011, 208, 1163–1177. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, N.C.; Lozier, A.; Flament, C.; Ricciardi-Castagnoli, P.; Bellet, D.; Suter, M.; Perricaudet, M.; Tursz, T.; Maraskovsky, E.; Zitvogel, L. Dendritic cells directly trigger NK cell functions: Cross-talk relevant in innate anti-tumor immune responses in vivo. Nat. Med. 1999, 5, 405–411. [Google Scholar] [CrossRef]
- Gottschalk, C.; Mettke, E.; Kurts, C. The Role of Invariant Natural Killer T Cells in Dendritic Cell Licensing, Cross-Priming, and Memory CD8(+) T Cell Generation. Front. Immunol. 2015, 6, 379. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, H.; Iwakabe, K.; Yahata, T.; Nishimura, S.; Ohta, A.; Ohmi, Y.; Sato, M.; Takeda, K.; Okumura, K.; Van Kaer, L.; et al. The natural killer T (NKT) cell ligand alpha-galactosylceramide demonstrates its immunopotentiating effect by inducing interleukin (IL)-12 production by dendritic cells and IL-12 receptor expression on NKT cells. J. Exp. Med. 1999, 189, 1121–1128. [Google Scholar] [CrossRef] [Green Version]
- Tahara, H.; Zitvogel, L.; Storkus, W.J.; Zeh, H.J., 3rd; McKinney, T.G.; Schreiber, R.D.; Gubler, U.; Robbins, P.D.; Lotze, M.T. Effective eradication of established murine tumors with IL-12 gene therapy using a polycistronic retroviral vector. J. Immunol. (Baltim. Md. 1950) 1995, 154, 6466–6474. [Google Scholar]
- Tatsumi, T.; Takehara, T.; Yamaguchi, S.; Sasakawa, A.; Miyagi, T.; Jinushi, M.; Sakamori, R.; Kohga, K.; Uemura, A.; Ohkawa, K.; et al. Injection of IL-12 gene-transduced dendritic cells into mouse liver tumor lesions activates both innate and acquired immunity. Gene Ther. 2007, 14, 863–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zitvogel, L.; Mayordomo, J.I.; Tjandrawan, T.; DeLeo, A.B.; Clarke, M.R.; Lotze, M.T.; Storkus, W.J. Therapy of murine tumors with tumor peptide-pulsed dendritic cells: Dependence on T cells, B7 costimulation, and T helper cell 1-associated cytokines. J. Exp. Med. 1996, 183, 87–97. [Google Scholar] [CrossRef]
- Cloosen, S.; Arnold, J.; Thio, M.; Bos, G.M.; Kyewski, B.; Germeraad, W.T. Expression of tumor-associated differentiation antigens, MUC1 glycoforms and CEA, in human thymic epithelial cells: Implications for self-tolerance and tumor therapy. Cancer Res. 2007, 67, 3919–3926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Jong, V.M.; Abreu, J.R.; Verrijn Stuart, A.A.; van der Slik, A.R.; Verhaeghen, K.; Engelse, M.A.; Blom, B.; Staal, F.J.; Gorus, F.K.; Roep, B.O. Alternative splicing and differential expression of the islet autoantigen IGRP between pancreas and thymus contributes to immunogenicity of pancreatic islets but not diabetogenicity in humans. Diabetologia 2013, 56, 2651–2658. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science (N. Y.) 2015, 348, 69–74. [Google Scholar] [CrossRef] [Green Version]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.P.; Simon, P.; Lower, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrors, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef]
- Yadav, M.; Jhunjhunwala, S.; Phung, Q.T.; Lupardus, P.; Tanguay, J.; Bumbaca, S.; Franci, C.; Cheung, T.K.; Fritsche, J.; Weinschenk, T.; et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014, 515, 572–576. [Google Scholar] [CrossRef]
- Kenter, G.G.; Welters, M.J.; Valentijn, A.R.; Lowik, M.J.; Berends-van der Meer, D.M.; Vloon, A.P.; Essahsah, F.; Fathers, L.M.; Offringa, R.; Drijfhout, J.W.; et al. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N. Engl. J. Med. 2009, 361, 1838–1847. [Google Scholar] [CrossRef] [Green Version]
- Ott, P.A.; Hu, Z.; Keskin, D.B.; Shukla, S.A.; Sun, J.; Bozym, D.J.; Zhang, W.; Luoma, A.; Giobbie-Hurder, A.; Peter, L.; et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017, 547, 217–221. [Google Scholar] [CrossRef]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haslam, A.; Prasad, V. Estimation of the Percentage of US Patients With Cancer Who Are Eligible for and Respond to Checkpoint Inhibitor Immunotherapy Drugs. JAMA Netw. Open 2019, 2, e192535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gubin, M.M.; Zhang, X.; Schuster, H.; Caron, E.; Ward, J.P.; Noguchi, T.; Ivanova, Y.; Hundal, J.; Arthur, C.D.; Krebber, W.J.; et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014, 515, 577–581. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- McGranahan, N.; Furness, A.J.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T.; et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science (N. Y.) 2016, 351, 1463–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (N. Y.) 2015, 348, 124–128. [Google Scholar] [CrossRef] [Green Version]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science (N. Y.) 2015, 350, 207–211. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Karaki, S.; Anson, M.; Tran, T.; Giusti, D.; Blanc, C.; Oudard, S.; Tartour, E. Is There Still Room for Cancer Vaccines at the Era of Checkpoint Inhibitors. Vaccines 2016, 4, 37. [Google Scholar] [CrossRef] [Green Version]
- Kather, J.N.; Suarez-Carmona, M.; Charoentong, P.; Weis, C.A.; Hirsch, D.; Bankhead, P.; Horning, M.; Ferber, D.; Kel, I.; Herpel, E.; et al. Topography of cancer-associated immune cells in human solid tumors. eLife 2018, 7, e36967. [Google Scholar] [CrossRef] [PubMed]
- Heger, L.; Amon, L.; Lehmann, C.H.K.; Dudziak, D. Systems Immunology Approaches for Understanding of Primary Dendritic Cell Subpopulations in the Past, Presence and Future. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar] [CrossRef]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amon, L.; Hatscher, L.; Heger, L.; Dudziak, D.; Lehmann, C.H.K. Harnessing the Complete Repertoire of Conventional Dendritic Cell Functions for Cancer Immunotherapy. Pharmaceutics 2020, 12, 663. https://doi.org/10.3390/pharmaceutics12070663
Amon L, Hatscher L, Heger L, Dudziak D, Lehmann CHK. Harnessing the Complete Repertoire of Conventional Dendritic Cell Functions for Cancer Immunotherapy. Pharmaceutics. 2020; 12(7):663. https://doi.org/10.3390/pharmaceutics12070663
Chicago/Turabian StyleAmon, Lukas, Lukas Hatscher, Lukas Heger, Diana Dudziak, and Christian H. K. Lehmann. 2020. "Harnessing the Complete Repertoire of Conventional Dendritic Cell Functions for Cancer Immunotherapy" Pharmaceutics 12, no. 7: 663. https://doi.org/10.3390/pharmaceutics12070663