Controlled Release of Doxorubicin from the Drug Delivery Formulation Composed of Single-Walled Carbon Nanotubes and Congo Red: A Molecular Dynamics Study and Dynamic Light Scattering Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Preparation of SWNT-CR and SWNT-CR-DOX Complexes for DLS Analysis
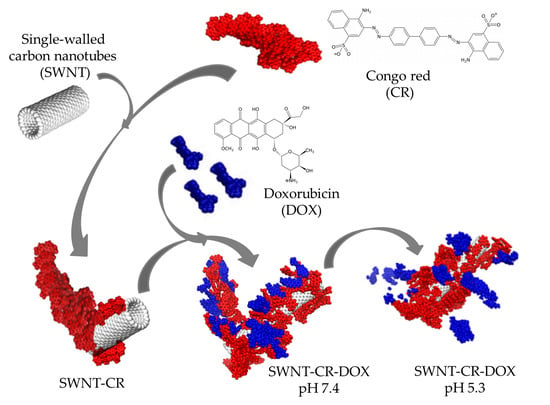
Formation of SWNT-CR Complexes
Formation of SWNT-CR-DOX Complexes
2.2.2. Dynamic Light Scattering (DLS)
2.2.3. Scanning Electron Microscopy (SEM)
2.2.4. Molecular Dynamics (MD)
3. Results and Discussion
3.1. The Effect of pH on DOX Release from a Triple SWNT-CR-DOX Complex (DLS Analysis)
3.2. Co-Adsorption of CR with DOX on SWNT (MD Analysis)
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| aiREBO | adaptive intermolecular reactive empirical bond order potential, |
| AMBER | assisted model building with energy refinement, |
| CHARMM | chemistry at Harvard macromolecular mechanics, |
| CR | Congo red, |
| CR-DOX | Congo red complexed with doxorubicin, |
| DLS | dynamic light scattering, |
| DOX | doxorubicin, |
| GAFF | generalized AMBER force field, |
| LAMMPS | open source code (Large-scale Atomic/ Molecular Massively Parallel Simulator), |
| MD | molecular dynamics, |
| RDF | radial distribution function, |
| RESP | restrained electrostatic potential, |
| SEM | scanning electron microscopy, |
| SRLS | self-assembled ribbon-like structures, |
| SWNT | single walled carbon nanotubes, carboxylic acid functionalized, |
| SWNT-CR | single walled carbon nanotubes, carboxylic acid functionalized complexed with Congo red, |
| SWNT-CR-DOX | single walled carbon nanotubes, carboxylic acid functionalized complexed with Congo red and doxorubicin. |
References
- Chaban, V.V.; Prezhdo, O.V. Water boiling inside carbon nanotubes: Toward efficient drug release. ACS Nano 2011, 5, 5647–5655. [Google Scholar] [CrossRef]
- Chytil, P.; Koziolová, E.; Etrych, T.; Ulbrich, K. HPMA Copolymer-Drug Conjugates with Controlled Tumor-Specific Drug Release. Macromol. Biosci. 2018, 18, 1700209. [Google Scholar] [CrossRef]
- Mikac, U.; Sepe, A.; Baumgartner, S.; Kristl, J. The Influence of High Drug Loading in Xanthan Tablets and Media with Different Physiological pH and Ionic Strength on Swelling and Release. Mol. Pharm. 2016, 13, 1147–1157. [Google Scholar] [CrossRef]
- Cheng, L.; Gao, S.; Ouyang, D.; Wang, H.; Wang, Y.; Pan, W.; Yang, X. Release Mechanism Between Ion Osmotic Pressure and Drug Release in Ionic-Driven Osmotic Pump Tablets (I). AAPS PharmSciTech. 2018, 19, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, X.; Zhou, Z.; Wang, K.; Li, C.; Qiao, H.; Oupicky, D.; Sun, M. Near-infrared light-triggered drug release from a multiple lipid carrier complex using an all-in-one strategy. J. Control. Release 2017, 261, 126–137. [Google Scholar] [CrossRef]
- Guha, S.; Shaw, S.K.; Spence, G.T.; Roland, F.M.; Smith, B.D. Clean Photothermal Heating and Controlled Release from Near-Infrared Dye Doped Nanoparticles without Oxygen Photosensitization. Langmuir 2015, 31, 7826–7834. [Google Scholar] [CrossRef] [Green Version]
- Panczyk, T.; Warzocha, T.P. Monte Carlo Study of the Properties of a Carbon Nanotube Functionalized by Magnetic Nanoparticles. J. Phys. Chem. C 2009, 113, 19155–19160. [Google Scholar] [CrossRef]
- Byrne, D.J.; Betancourt, T.; Brannon-Peppas, L. Active targeting schemes for nanoparticle systems in cancer therapeutics. Adv. Drug Deliv. Rev. 2008, 60, 1615–1626. [Google Scholar] [CrossRef]
- Zhu, J.; Liao, L.; Bian, X.; Kong, J.; Yang, P.; Liu, B. pH-controlled delivery of doxorubicin to cancer cells, based on small mesoporous carbon nanospheres. Small 2012, 8, 2715–2720. [Google Scholar] [CrossRef]
- Kato, Y.; Ozawa, S.; Miyamoto, C.; Maehata, Y.; Suzuki, A.; Maeda, T.; Baba, Y. Acidic Extracellular microenvironment and cancer. Cancer Cell Int. 2013, 13, 89. [Google Scholar] [CrossRef] [Green Version]
- Hakeem, A.; Zhan, G.; Xu, Q.; Yong, T.; Yang, X.; Gan, L. Facile synthesis of pH-responsive doxorubicin-loaded layered double hydroxide for efficient cancer therapy. J. Mater. Chem. B. 2018, 6, 5768–5774. [Google Scholar] [CrossRef]
- Jagusiak, A.; Chłopaś, K.; Zemanek, G.; Jemioła-Rzemińska, M.; Piekarska, B.; Stopa, B.; Panczyk, T. Self-Assembled Supramolecular Ribbon-Like Structures Complexed to Single Walled Carbon Nanotubes as Possible Anticancer Drug Delivery Systems. Int. J. Mol. Sci. 2019, 20, 2064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rybarska, J.; Piekarska, B.; Stopa, B.; Zemanek, G.; Konieczny, L.; Roterman, I. Chapter: Supramolecular systems as protein ligands. In Self-Assembled Molecules—New Kind of Protein Ligands: Supramolecular Ligands, 1st ed.; Roterman, I., Konieczny, L., Eds.; Springer International Publishing: New York, NY, USA, 2008; Volume 1, pp. 1–20. [Google Scholar]
- Jagusiak, A. An outline of the use of supramolecular compounds in biology and medicine. Acta Biochim. Pol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Piekarska, B.; Konieczny, L.; Rybarska, J.; Stopa, B.; Zemanek, G.; Szneler, E.; Król, M.; Nowak, M.; Roterman, I. Heat-induced formation of a specific binding site for self-assembled Congo red in the V domain of immunoglobulin L chain λ. Biopolymers 2001, 59, 446–456. [Google Scholar] [CrossRef]
- Piekarska, B.; Konieczny, L.; Rybarska, J.; Stopa, B.; Spólnik, P.; Roterman, I.; Król, M. Intramolecular signaling in immunoglobulins—New evidence emerging from the use of supramolecular protein ligands. J. Physiol. Pharmacol. 2004, 55, 487–501. [Google Scholar]
- Jagusiak, A.; Rybarska, J.; Piekarska, B.; Stopa, B.; Konieczny, L. Chapter: Supramolecular Congo red as specific ligand of antibodies engaged in immune complex. In Self-Assembled Molecules—New Kind of Protein Ligands: Supramolecular Ligands, 1st ed.; Roterman, I., Konieczny, L., Eds.; Springer International Publishing: New York, NY, USA, 2008; Volume 1, pp. 21–42. [Google Scholar]
- Koshland, D.E., Jr. Enzyme flexibility and enzyme action. J. Cell. Comp. Physiol. 1959, 54, 245–258. [Google Scholar] [CrossRef]
- Brown, M.A.; Mitar, D.A.; Whitworth, J.A. Measurement of Plasma Volume in Pregnancy. Clin. Sci. 1992, 83, 29–34. [Google Scholar] [CrossRef]
- Jagusiak, A.; Panczyk, T. Interaction of Congo Red, Evans Blue and Titan Yellow with doxorubicin in aqueous solutions. A molecular dynamics study. J. Mol. Liq. 2019, 279, 640–648. [Google Scholar] [CrossRef]
- Prato, M.; Kostarelos, K.; Bianco, A. Functionalized carbon nanotubes in drug design and discovery. Acc. Chem. Res. 2008, 41, 60–68. [Google Scholar] [CrossRef]
- Pastorin, G. Carbon Nanotubes: From Bench Chemistry to Promising Biomedical Applications; Pastorin, G., Ed.; Pan Stanford Publishing: Singapore, 2011; ISBN1 9814241660. ISBN2 9789814241663. [Google Scholar]
- Wong, B.S.; Yoong, S.L.; Jagusiak, A.; Panczyk, T. Carbon nanotubes for delivery of small molecule drugs. Adv. Drug Deliv. Rev. 2013, 65, 1964–2015. [Google Scholar] [CrossRef]
- Kamalha, E.; Shi, X.; Mwasiagi, J.; Zeng, Y. Nanotechnology and carbon nanotubes; a review of potential in drug delivery. Macromol. Res. 2012, 20, 891–898. [Google Scholar] [CrossRef]
- Rastogi, V.; Yadav, P.; Bhattacharya, S.S.; Mishra, A.K.; Verma, N.; Verma, A.; Pandit, J.K. Carbon nanotubes: An emerging drug carrier for targeting cancer cells. J. Drug Deliv. 2014, 2014, 670815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamon, M.A.; Chen, J.; Hu, H.; Chen, Y.; Itkis, M.E.; Rao, A.M.; Eklund, P.C.; Haddon, R.C. Dissolution of single-walled carbon nanotubes. Adv. Mater. 1999, 11, 834–840. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, X.; Nakayama-Ratchford, N.; Dai, H.J. Supramolecular chemistry on water-soluble carbon nanotubes for drug loading and delivery. ASC Nano 2007, 1, 50–56. [Google Scholar] [CrossRef]
- Tasis, D.; Tagmatarchis, N.; Bianco, A.; Prato, M. Chemistry of carbon nanotubes. Chem. Rev. 2006, 106, 1105–1136. [Google Scholar] [CrossRef]
- Nepal, D.; Geckeler, K.E. Proteins and carbon nanotubes: Close encounter in water. Small 2007, 3, 1259–1265. [Google Scholar] [CrossRef]
- Witus, L.S.; Rocha, J.D.R.; Yuwono, V.M.; Paramanov, S.E.; Weisman, R.B.; Hartgerink, J.D. Peptides that non-covalently functionalize single-walled carbon nanotubes to give controlled solubility characteristics. J. Mater. Chem. 2007, 17, 1909–1915. [Google Scholar] [CrossRef]
- Ciofani, J.; Obata, Y.; Sato, I.; Okamura, Y.; Raffa, V.; Manciassi, A.; Dario, P.; Takeda, N.; Takeoka, S. Realization, characterization and functionalization of lipidic wrapped carbon nanotubes. J. Nanopart. Res. 2009, 11, 477–484. [Google Scholar] [CrossRef]
- Chhikara, B.S.; Misra, S.K.; Bhattacharya, S. CNT loading into cationic cholesterol suspensions show improved DNA binding and serum stability and ability to internalize into cancer cells. Nanotechnology 2012, 23, 065101. [Google Scholar] [CrossRef]
- Zhang, X.; Meng, L.; Lu, Q.; Fei, Z.; Dyson, P.J. Targeted delivery and controlled release of doxorubicin to cancer cells using modified single wall carbon nanotubes. Biomaterials 2009, 30, 6041–6047. [Google Scholar] [CrossRef]
- Ajima, K.; Murakami, T.; Mizoguchi, Y.; Tsuchida, K.; Ichihashi, T.; Iijima, S.; Yudasaka, M. Enhancement of in vivo anticancer effects of cisplatin by incorporation inside single-wall carbon nanohorns. ACS Nano 2008, 2, 2057–2064. [Google Scholar] [CrossRef]
- Ren, Y.P.; Pastorin, G. Incorporation of hexamethylmelamine inside capped carbon nanotubes. Adv. Mater. 2008, 20, 2031–2036. [Google Scholar] [CrossRef]
- Luo, X.; Matranga, C.; Tan, S.; Alba, N.; Cui, X.T. Carbon nanotube nanoreservoir for controlled release of anti-inflammatory dexamethasone. Biomaterials 2011, 32, 6316–6323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagusiak, A.; Piekarska, B.; Panczyk, T.; Jemiolarzeminska, M.; Bielanska, E.; Stopa, B.; Zemanek, G.; Rybarska, J.; Roterman, I.; Konieczny, L.; et al. Dispersion of single-wall carbon nanotubes with supramolecular Congo red—Properties of the complexes and mechanism of the interaction. Beilstein J. Nanotechnol. 2017, 8, 636–648. [Google Scholar] [CrossRef] [Green Version]
- Panczyk, T.; Wolski, P.; Jagusiak, A.; Drach, M. Molecular dynamics study of Congo red interactionwith carbon nanotubes. RSC Adv. 2014, 4, 47304–47312. [Google Scholar] [CrossRef]
- Król, M.; Borowski, T.; Roterman, I.; Piekarska, B.; Stopa, B.; Rybarska, J.; Konieczny, L. Force-field parametrization and molecular dynamics simulations of Congo red. J. Comput. Aided Mol. Des. 2004, 18, 41–53. [Google Scholar] [CrossRef]
- Panczyk, T.; Wolski, P.; Lajtar, L. Coadsorption of Doxorubicin and Selected Dyes on Carbon Nanotubes. Theoretical Investigation of Potential Application as a pH-Controlled Drug Delivery System. Langmuir 2016, 32, 4719–4728. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amberforce field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Bayly, C.I.; Cieplak, P.; Cornell, W.; Kollman, P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Wu, C.; Wang, Z.; Lei, H.; Zhang, W.; Duan, Y. Dual Binding Modes of Congo Red to Amyloid Protofibril Surface Observed in Molecular Dynamics Simulations. J. Am. Chem. Soc. 2007, 129, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Dupradeaum, F.; Pigache, A.; Zaffran, T.; Savineau, C.; Lelong, R.; Grivel, N.; Lelong, D.; Rosanski, W.; Cieplak, P. The R.E.D. tools: Advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010, 12, 7821–7839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wikipedia, the Free Encyclopedia. Available online: http://en.wikipedia.org/wiki/Water_model (accessed on 8 January 2014).
- Stuart, S.J.; Tutein, A.B.; Harrison, J.A. A reactive potential for hydrocarbons with intermolecular interactions. J. Chem. Phys. 2000, 112, 6472. [Google Scholar] [CrossRef] [Green Version]
- Plimpton, S. Fast Parallel Algorithms for Short-Range Molecular Dynamics. J. Comput. Phys. 1995, 117, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Fülöp, Z.; Gref, R.; Loftsson, T. A permeation method for detection of self-aggregation of doxorubicin in aqueous environment. Int. J. Pharm. 2013, 454, 559–561. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.J. DLS and zeta potential–What they are and what they are not? J. Cont. Release 2016, 235, 337–351. [Google Scholar] [CrossRef] [PubMed]















| Formulation Energy kJ/mol | SWNT (10.0) + 20CR + 10DOX | SWNT (10.0) + 50CR + 20DOX | SWNT (30.0) + 20CR + 10DOX | SWNT (30.0) + 40CR + 10DOX |
|---|---|---|---|---|
| Neutral pH | ||||
| UCR-CR | 486 | 1010 | 278 | 451 |
| UCR-SWNT | −87 | −57 | −216 | −198 |
| UDOX-DOX | −731 | −700 | −788 | −786 |
| UDOX-SWNT | −46 | −10 | −152 | −168 |
| UCR-DOX | −36 | −20 | −12 | −9 |
| Intermediate pH (between Neutral and Acidic pH) | ||||
| UCR-CR | −172 | - | −236 | −241 |
| UCR-SWNT | −74 | - | −207 | −119 |
| UDOX-DOX | −748 | - | −773 | −800 |
| UDOX-SWNT | −35 | - | −142 | −156 |
| UCR-DOX | −27 | - | −7 | −7 |
| Acidic pH | ||||
| UCR-CR | −703 | −720 | −646 | −647 |
| UCR- SWNT | −48 | −51 | −201 | −141 |
| UDOX-DOX | −803 | −757 | −776 | −777 |
| UDOX-SWNT | −53 | 0 | −180 | −186 |
| UCR-DOX | −5 | −2 | −0.2 | -2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jagusiak, A.; Chlopas, K.; Zemanek, G.; Wolski, P.; Panczyk, T. Controlled Release of Doxorubicin from the Drug Delivery Formulation Composed of Single-Walled Carbon Nanotubes and Congo Red: A Molecular Dynamics Study and Dynamic Light Scattering Analysis. Pharmaceutics 2020, 12, 622. https://doi.org/10.3390/pharmaceutics12070622
Jagusiak A, Chlopas K, Zemanek G, Wolski P, Panczyk T. Controlled Release of Doxorubicin from the Drug Delivery Formulation Composed of Single-Walled Carbon Nanotubes and Congo Red: A Molecular Dynamics Study and Dynamic Light Scattering Analysis. Pharmaceutics. 2020; 12(7):622. https://doi.org/10.3390/pharmaceutics12070622
Chicago/Turabian StyleJagusiak, Anna, Katarzyna Chlopas, Grzegorz Zemanek, Pawel Wolski, and Tomasz Panczyk. 2020. "Controlled Release of Doxorubicin from the Drug Delivery Formulation Composed of Single-Walled Carbon Nanotubes and Congo Red: A Molecular Dynamics Study and Dynamic Light Scattering Analysis" Pharmaceutics 12, no. 7: 622. https://doi.org/10.3390/pharmaceutics12070622