Adeno-Associated Virus Serotype 8-Mediated Genetic Labeling of Cholangiocytes in the Neonatal Murine Liver
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Experiment
2.2. Immunofluorescence
2.3. Quantification and Data Analysis
3. Results
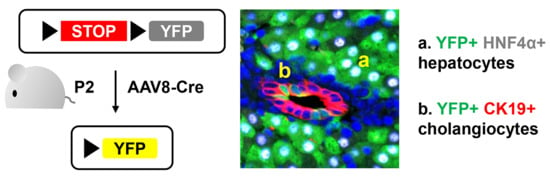
3.1. Neonatal Injection of AAV8-CMV-Cre Labels Cholangiocytes
3.2. AAV8-CMV-Cre Labels CD34-Expressing Cells
3.3. Neonatal Injection of AAV8-TBG-Cre Labels Cholangiocytes
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, Y.; Fang, B.; Damle, M.; Guan, D.; Li, Z.; Kim, Y.H.; Gannon, M.; Lazar, M.A. HNF6 and Rev-erbalpha integrate hepatic lipid metabolism by overlapping and distinct transcriptional mechanisms. Genes Dev. 2016, 30, 1636–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Kim, K.; Wang, X.; Bartolome, A.; Salomao, M.; Dongiovanni, P.; Meroni, M.; Graham, M.J.; Yates, K.P.; Diehl, A.M.; et al. Hepatocyte Notch activation induces liver fibrosis in nonalcoholic steatohepatitis. Sci. Transl. Med. 2018, 10, eaat0344. [Google Scholar] [CrossRef] [PubMed]
- Morell, C.M.; Fiorotto, R.; Meroni, M.; Raizner, A.; Torsello, B.; Cadamuro, M.; Spagnuolo, G.; Kaffe, E.; Sutti, S.; Albano, E.; et al. Notch signaling and progenitor/ductular reaction in steatohepatitis. PLoS ONE 2017, 12, e0187384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, A.; Magee, N.; Deng, F.; Lehn, S.; Zhong, C.; Zhang, Y. Hepatocyte nuclear receptor SHP suppresses inflammation and fibrosis in a mouse model of nonalcoholic steatohepatitis. J. Biol. Chem. 2018, 293, 8656–8671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walesky, C.; Gunewardena, S.; Terwilliger, E.F.; Edwards, G.; Borude, P.; Apte, U. Hepatocyte-specific deletion of hepatocyte nuclear factor-4alpha in adult mice results in increased hepatocyte proliferation. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G26–G37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, X.; Espanol-Suner, R.; Mederacke, I.; Affo, S.; Manco, R.; Sempoux, C.; Lemaigre, F.P.; Adili, A.; Yuan, D.; Weber, A.; et al. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. J. Clin. Investig. 2015, 125, 3891–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherman, A.; Biswas, M.; Herzog, R.W. Innovative Approaches for Immune Tolerance to Factor VIII in the Treatment of Hemophilia A. Front. Immunol. 2017, 8, 1604. [Google Scholar] [CrossRef] [Green Version]
- Zaiss, A.K.; Muruve, D.A. Immune responses to adeno-associated virus vectors. Curr. Gene Ther. 2005, 5, 323–331. [Google Scholar] [CrossRef]
- Bortolussi, G.; Zentillin, L.; Vanikova, J.; Bockor, L.; Bellarosa, C.; Mancarella, A.; Vianello, E.; Tiribelli, C.; Giacca, M.; Vitek, L.; et al. Life-long correction of hyperbilirubinemia with a neonatal liver-specific AAV-mediated gene transfer in a lethal mouse model of Crigler-Najjar Syndrome. Hum. Gene Ther. 2014, 25, 844–855. [Google Scholar] [CrossRef] [Green Version]
- Sabatino, D.E.; Mackenzie, T.C.; Peranteau, W.; Edmonson, S.; Campagnoli, C.; Liu, Y.L.; Flake, A.W.; High, K.A. Persistent expression of hF.IX After tolerance induction by in utero or neonatal administration of AAV-1-F.IX in hemophilia B mice. Mol. Ther. 2007, 15, 1677–1685. [Google Scholar] [CrossRef]
- Wang, L.; Wang, H.; Bell, P.; McMenamin, D.; Wilson, J.M. Hepatic gene transfer in neonatal mice by adeno-associated virus serotype 8 vector. Hum. Gene Ther. 2012, 23, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Calcedo, R.; Morizono, H.; Wang, L.; McCarter, R.; He, J.; Jones, D.; Batshaw, M.L.; Wilson, J.M. Adeno-associated virus antibody profiles in newborns, children, and adolescents. Clin. Vaccine Immunol. 2011, 18, 1586–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Wang, L.; Bell, P.; McMenamin, D.; He, Z.; White, J.; Yu, H.; Xu, C.; Morizono, H.; Musunuru, K.; et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016, 34, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, P.; Rosario, A.; Cruz, P.; Siemienski, Z.; Ceballos-Diaz, C.; Crosby, K.; Jansen, K.; Borchelt, D.R.; Kim, J.Y.; Jankowsky, J.L.; et al. Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS ONE 2013, 8, e67680. [Google Scholar] [CrossRef]
- Colella, P.; Ronzitti, G.; Mingozzi, F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2018, 8, 87–104. [Google Scholar] [CrossRef] [Green Version]
- Nishio, T.; Hu, R.; Koyama, Y.; Liang, S.; Rosenthal, S.B.; Yamamoto, G.; Karin, D.; Baglieri, J.; Ma, H.Y.; Xu, J.; et al. Activated hepatic stellate cells and portal fibroblasts contribute to cholestatic liver fibrosis in MDR2 knockout mice. J. Hepatol. 2019, 71, 573–585. [Google Scholar] [CrossRef]
- Park, S.M. The crucial role of cholangiocytes in cholangiopathies. Gut Liver 2012, 6, 295–304. [Google Scholar] [CrossRef]
- Peters, K.M.; Wilson, R.B.; Borradaile, N.M. Non-parenchymal hepatic cell lipotoxicity and the coordinated progression of non-alcoholic fatty liver disease and atherosclerosis. Curr. Opin. Lipidol. 2018, 29, 417–422. [Google Scholar] [CrossRef]
- Seo, W.; Jeong, W.I. Hepatic non-parenchymal cells: Master regulators of alcoholic liver disease? World J. Gastroenterol. 2016, 22, 1348–1356. [Google Scholar] [CrossRef]
- Wang, P.; Koyama, Y.; Liu, X.; Xu, J.; Ma, H.Y.; Liang, S.; Kim, I.H.; Brenner, D.A.; Kisseleva, T. Promising Therapy Candidates for Liver Fibrosis. Front Physiol. 2016, 7, 47. [Google Scholar] [CrossRef] [Green Version]
- Powell, S.K.; Rivera-Soto, R.; Gray, S.J. Viral expression cassette elements to enhance transgene target specificity and expression in gene therapy. Discov. Med. 2015, 19, 49–57. [Google Scholar] [PubMed]
- Srinivas, S.; Watanabe, T.; Lin, C.S.; William, C.M.; Tanabe, Y.; Jessell, T.M.; Costantini, F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001, 1, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwai, H.; Hoberman, A.M. Oral (Gavage) Combined Developmental and Perinatal/Postnatal Reproduction Toxicity Study of Ammonium Salt of Perfluorinated Hexanoic Acid in Mice. Int. J. Toxicol. 2014, 33, 219–237. [Google Scholar] [CrossRef] [PubMed]
- Higashi, S.; Hioki, K.; Kurotani, T.; Kasim, N.; Molnar, Z. Functional thalamocortical synapse reorganization from subplate to layer IV during postnatal development in the reeler-like mutant rat (shaking rat Kawasaki). J. Neurosci. 2005, 25, 1395–1406. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Tanimizu, N.; Nakamura, Y.; Ichinohe, N.; Mizuguchi, T.; Hirata, K.; Mitaka, T. Hepatic biliary epithelial cells acquire epithelial integrity but lose plasticity to differentiate into hepatocytes in vitro during development. J. Cell Sci. 2013, 126, 5239–5246. [Google Scholar] [CrossRef] [Green Version]
- Poncy, A.; Antoniou, A.; Cordi, S.; Pierreux, C.E.; Jacquemin, P.; Lemaigre, F.P. Transcription factors SOX4 and SOX9 cooperatively control development of bile ducts. Dev. Biol. 2015, 404, 136–148. [Google Scholar] [CrossRef]
- Lesaffer, B.; Verboven, E.; Van Huffel, L.; Moya, I.M.; van Grunsven, L.A.; Leclercq, I.A.; Lemaigre, F.P.; Halder, G. Comparison of the Opn-CreER and Ck19-CreER Drivers in Bile Ducts of Normal and Injured Mouse Livers. Cells 2019, 8, 380. [Google Scholar] [CrossRef] [Green Version]
- Nikolic, I.; Todorovic, V.; Petrovic, A.; Petrovic, V.; Jovic, M.; Vladicic, J.; Puskas, N. Immunohistochemical Heterogeneity of the Endothelium of Blood and Lymphatic Vessels in the Developing Human Liver and in Adulthood. Cells Tissues Organs 2017, 203, 243–257. [Google Scholar] [CrossRef]
- Wang, L.; Wang, H.; Bell, P.; McCarter, R.J.; He, J.; Calcedo, R.; Vandenberghe, L.H.; Morizono, H.; Batshaw, M.L.; Wilson, J.M. Systematic evaluation of AAV vectors for liver directed gene transfer in murine models. Mol. Ther. 2010, 18, 118–125. [Google Scholar] [CrossRef]






| Target | Host Species | Source (Catalog Number) | Dilution |
|---|---|---|---|
| YFP | Goat | Abcam (ab6673) | 250 |
| YFP | Chicken | Aves Labs (GFP-1020) | 500 |
| CK19 | Rabbit | Abcam (ab52625) | 200 |
| EPCAM | Rabbit | Abcam (ab71916) | 100 |
| OPN | Goat | R&D Systems (AF808) | 200 |
| HNF4α | Mouse | R&D Systems (PP-H1415-00) | 400 |
| CD34 | Rabbit | Abcam (ab81289) | 100 |
| αSMA | Mouse | Abcam (ab7817) | 200 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Zhou, P.; Whyte, S.; Shin, S. Adeno-Associated Virus Serotype 8-Mediated Genetic Labeling of Cholangiocytes in the Neonatal Murine Liver. Pharmaceutics 2020, 12, 351. https://doi.org/10.3390/pharmaceutics12040351
Lee S, Zhou P, Whyte S, Shin S. Adeno-Associated Virus Serotype 8-Mediated Genetic Labeling of Cholangiocytes in the Neonatal Murine Liver. Pharmaceutics. 2020; 12(4):351. https://doi.org/10.3390/pharmaceutics12040351
Chicago/Turabian StyleLee, Sanghoon, Ping Zhou, Senyo Whyte, and Soona Shin. 2020. "Adeno-Associated Virus Serotype 8-Mediated Genetic Labeling of Cholangiocytes in the Neonatal Murine Liver" Pharmaceutics 12, no. 4: 351. https://doi.org/10.3390/pharmaceutics12040351