Nanovesicle-Mediated Delivery Systems for CRISPR/Cas Genome Editing
Abstract
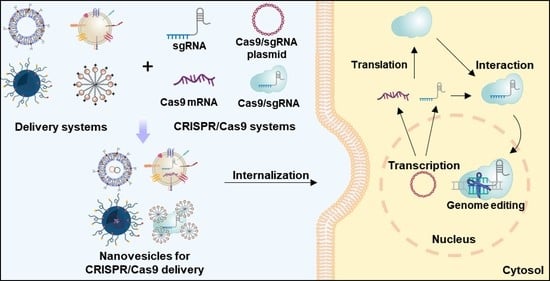
:1. Introduction
2. Nonviral Delivery Systems for Genome Editing
2.1. Lipid-Based Vesicles for Genome Editing
2.1.1. Cationic Lipid Nanoparticles
2.1.2. Lipid Nanoshells
2.2. Polymer-Based Delivery Systems
2.2.1. Polyethylenimine
2.2.2. Polyamidoamine
2.2.3. Other Synthetic Polymers
2.2.4. Chitosan
2.3. Extracellular Vesicles
2.3.1. Exosomes
2.3.2. Microvesicles
2.4. Peptide/Protein-Based Systems
2.4.1. Peptides
2.4.2. Proteins
2.4.3. Virus-Like Particles
3. Targeting Ligands for Genome Editing
3.1. Chemical Ligands for Targeted Delivery
3.2. Peptide Ligands for Targeted Delivery
3.3. Antibodies and Aptamers for Targeted Delivery
4. Delivery Strategies for Genome Editing
4.1. Chemical/Molecular Delivery Strategies
4.1.1. Chemical/Peptide-Enhanced Delivery
4.1.2. Light-Enhanced Delivery
4.1.3. Glutathione-Triggered Delivery
4.1.4. pH-Responsive Delivery
4.2. Physical Delivery Strategies
5. Challenges and Future Perspectives
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Abbreviation | Full Name |
| ANGPTL3 | Angiopoietin like 3 |
| APOC3 | Apolipoprotein C |
| ASGP | Asialoglycoprotein |
| Cas | Clustered regularly interspaced short palindromic repeats-associated protein |
| CDC6 | Cell division control protein 6 |
| C/EBPα | CCAAT enhancer binding protein alpha |
| Chol-PEG | Polyethylene glycol-linked cholesterol |
| CPP | Cell-penetrating peptide |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| crRNAs | CRISPR RNAs |
| CTNNB1 | β-catenin |
| DDX3 | DEAD-box RNA helicase 3 |
| DNMT1 | DNA-methyltransferase 1 |
| DOPE | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine |
| DOTAP | 1,2-dioleoyl-3-trimethylammoniumpropane |
| DSPE-PEG-2000 | 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethyleneglycol)-2000] |
| EGFP | Enhanced green fluorescent protein |
| EMX1 | Empty spiracles homeobox 1 |
| FKBP | FK506-binding protein |
| FRB | FKBP-rapamycin binding domain |
| GFP | Green fluorescent protein |
| GPX4 | Glutathione peroxidase-4 |
| GSH | Glutathione |
| HPD | Hydroxyphenylpyruvate dioxygenase |
| HPV16 E6/E7 | Human papillomavirus 16 E6/E7 |
| ICAM1 | Intercellular adhesion molecule 1 |
| IDUA | α-L-iduronidase |
| IQGAP1 | IQ motif-containing GTPase-activating protein 1 |
| miRNA | MicroRNA |
| MTH1 | MutT homolog 1 |
| NIR | Near-infrared |
| Nrf2 | Nuclear factor erythroid-2-related factor 2 |
| PAMAM | Polyamidoamine |
| PARP1 | Poly (ADP-ribose) polymerase-1 |
| pCas9 | Plasmid DNA encoding clustered regularly interspaced short palindromic Repeats-associated protein 9 |
| PCSK9 | Protease proprotein convertase subtilisin/kexin type 9 |
| PD-L1 | Programmed death-ligand 1 |
| PEG | Polyethylene glycol |
| PEI | Polyethyleneimine |
| PLK1 | Polo-like kinase 1 |
| PPABLG | Poly(γ-4-((2-(piperidin-1-yl)ethyl)aminomethyl)benzyl-L-glutamate) |
| psgRNA | Plasmid DNA encoding single-guide RNA |
| RHBDF1 | Rhomboid 5 homolog 1 |
| ROS | Reactive oxygen species |
| RUNX1 | Runt-related transcription factor 1 |
| sgRNA | Single-guide RNA |
| siRNA | Small inhibitory RNA |
| STAT3 | Signal transducer and activator of transcription 3 |
| STING | Stimulator of IFN genes |
| TALENs | Transcription activator-like effector nucleases |
| tracrRNAs | Trans-activating CRISPR RNAs |
| TYR | Tyrosinase |
| VEGF-A | Vascular endothelial growth factor A |
| VEGFR2 | Vascular endothelial growth factor receptor 2 |
| VLP | Virus-like particle |
| VSV | Vesicular stomatitis virus |
| ZFN | Zinc-finger nuclease |
| β-CD | β-cyclodextrin |
References
- Naldini, L. Gene therapy returns to centre stage. Nature 2015, 526, 351–360. [Google Scholar] [CrossRef]
- Cavazzana, M.; Bushman, F.D.; Miccio, A.; André-Schmutz, I.; Six, E. Gene therapy targeting haematopoietic stem cells for inherited diseases: Progress and challenges. Nat. Rev. Drug Discov. 2019, 18, 447–462. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Ren, Y.X.; Xie, Y.; Lu, W.L. Gene regulations and delivery vectors for treatment of cancer. J. Pharm. Investig. 2020, 50, 309–326. [Google Scholar] [CrossRef]
- CRICK, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Hyde, S.C.; Pringle, I.A.; Abdullah, S.; Lawton, A.E.; Davies, L.A.; Varathalingam, A.; Nunez-Alonso, G.; Green, A.M.; Bazzani, R.P.; Sumner-Jones, S.G.; et al. CpG-free plasmids confer reduced inflammation and sustained pulmonary gene expression. Nat. Biotechnol. 2008, 26, 549–551. [Google Scholar] [CrossRef]
- Hajj, K.A.; Whitehead, K.A. Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017, 2, 1–17. [Google Scholar] [CrossRef]
- Lee, J.; Cho, Y.J.; Lee, J.-W.; Ahn, H.J. KSP siRNA/paclitaxel-loaded PEGylated cationic liposomes for overcoming resistance to KSP inhibitors: Synergistic antitumor effects in drug-resistant ovarian cancer. J. Control. Release 2020, 321, 184–197. [Google Scholar] [CrossRef]
- Shim, G.; Choi, H.; Lee, S.; Choi, J.; Yu, Y.H.; Park, D.-E.; Choi, Y.; Kim, C.-W.; Oh, Y.-K. Enhanced intrapulmonary delivery of anticancer sirna for lung cancer therapy using cationic ethylphosphocholine-based nanolipoplexes. Mol. Ther. 2013, 21, 816–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.W.L.; Paoletti, C.; Campisi, M.; Osaki, T.; Adriani, G.; Kamm, R.D.; Mattu, C.; Chiono, V. MicroRNA delivery through nanoparticles. J. Control. Release 2019, 313, 80–95. [Google Scholar] [CrossRef] [PubMed]
- Cehajic-Kapetanovic, J.; Xue, K.; de la Camara, C.M.F.; Nanda, A.; Davies, A.; Wood, L.J.; Salvetti, A.P.; Fischer, M.D.; Aylward, J.W.; Barnard, A.R.; et al. Initial results from a first-in-human gene therapy trial on X-linked retinitis pigmentosa caused by mutations in RPGR. Nat. Med. 2020, 26, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Kohn, D.B.; Booth, C.; Kang, E.M.; Pai, S.Y.; Shaw, K.L.; Santilli, G.; Armant, M.; Buckland, K.F.; Choi, U.; De Ravin, S.S.; et al. Lentiviral gene therapy for X-linked chronic granulomatous disease. Nat. Med. 2020, 26, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, B.; Amador, G.; Viswanatha, R.; Zirin, J.; Mohr, S.E.; Perrimon, N. CRISPR-based engineering of gene knockout cells by homology-directed insertion in polyploid Drosophila S2R+ cells. Nat. Protoc. 2020, 15, 3478–3498. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.; Sreekanth, V.; Cox, K.J.; Law, B.K.; Wagner, B.K.; Karp, J.M.; Choudhary, A. Engineering designer beta cells with a CRISPR-Cas9 conjugation platform. Nat. Commun. 2020, 11, 4043. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef] [Green Version]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Dreyer, A.K.; Hoffmann, D.; Lachmann, N.; Ackermann, M.; Steinemann, D.; Timm, B.; Siler, U.; Reichenbach, J.; Grez, M.; Moritz, T.; et al. TALEN-mediated functional correction of X-linked chronic granulomatous disease in patient-derived induced pluripotent stem cells. Biomaterials 2015, 69, 191–200. [Google Scholar] [CrossRef]
- Holt, N.; Wang, J.; Kim, K.; Friedman, G.; Wang, X.; Taupin, V.; Crooks, G.M.; Kohn, D.B.; Gregory, P.D.; Holmes, M.C.; et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010, 28, 839–847. [Google Scholar] [CrossRef] [Green Version]
- Ding, W.; Zhu, D.; Hu, Z.; Jiang, X.; Yu, L.; Wang, X.; Zhang, C.; Wang, L.; Ji, T.; Li, K.; et al. Zinc finger nucleases targeting the human papillomavirus E7 oncogene induce E7 disruption and a transformed Phenotype in HPV16/18-positive cervical cancer cells. Clin. Cancer Res. 2014, 20, 6495–6503. [Google Scholar] [CrossRef] [Green Version]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef] [PubMed]
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalem, O.; Sanjana, N.E.; Zhang, F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015, 16, 299–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.H. Genome-editing technologies: Concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol. Ther. Nucleic Acids 2019, 16, 326–334. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.S.; Bishop, E.S.; Zhang, R.; Yu, X.; Farina, E.M.; Yan, S.; Zhao, C.; Zeng, Z.; Shu, Y.; Wu, X.; et al. Adenovirus-mediated gene delivery: Potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017, 4, 43–63. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Garrigue, A.; Wang, G.P.; Soulier, J.; Lim, A.; Morillon, E.; Clappier, E.; Caccavelli, L.; Delabesse, E.; Beldjord, K.; et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Investig. 2008, 118, 3132–3142. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, H.; Colosi, P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010, 18, 80–86. [Google Scholar] [CrossRef]
- Elmowafy, E.M.; Tiboni, M.; Soliman, M.E. Biocompatibility, biodegradation and biomedical applications of poly(lactic acid)/poly(lactic co glycolic acid) micro and nanoparticles. J. Pharm. Investig. 2019, 49, 347–380. [Google Scholar] [CrossRef]
- Zhen, S.; Liu, Y.; Lu, J.; Tuo, X.; Yang, X.; Chen, H.; Chen, W.; Li, X. Human papillomavirus oncogene manipulation using clustered regularly interspersed short palindromic repeats/cas9 delivered by ph-sensitive cationic liposomes. Hum. Gene Ther. 2020, 31, 309–324. [Google Scholar] [CrossRef]
- Schuh, R.S.; Poletto, É.; Pasqualim, G.; Tavares, A.M.V.; Meyer, F.S.; Gonzalez, E.A.; Giugliani, R.; Matte, U.; Teixeira, H.F.; Baldo, G. In vivo genome editing of mucopolysaccharidosis I mice using the CRISPR/Cas9 system. J. Control. Release 2018, 288, 23–33. [Google Scholar] [CrossRef]
- Yin, H.; Song, C.Q.; Suresh, S.; Wu, Q.; Walsh, S.; Rhym, L.H.; Mintzer, E.; Bolukbasi, M.F.; Zhu, L.J.; Kauffman, K.; et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017, 35, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, P.; Feng, Q.; Wang, N.; Chen, Z.; Huang, Y.; Zheng, W.; Jiang, X. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. NPG Asia Mater. 2017, 9, e441. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, L.; Xie, Y.; Wang, N.; Tang, R.; Zheng, W.; Jiang, X. Genome editing for cancer therapy: Delivery of Cas9 protein/sgRNA plasmid via a gold nanocluster/lipid core–shell nanocarrier. Adv. Sci. 2017, 4, 1700175. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Wang, H.X.; Lao, Y.H.; Hu, H.; Vatan, N.; Guo, J.; Ho, T.C.; Huang, D.; Li, M.; Shao, D.; et al. A versatile nonviral delivery system for multiplex gene-editing in the liver. Adv. Mater. 2020, 32, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Song, L.; Liu, X.; Yang, X.; Li, X.; He, T.; Wang, N.; Yang, S.; Yu, C.; Yin, T.; et al. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano 2017, 11, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, Z.; Zhu, S.; He, L.; Fan, W.; Tang, W.; Zou, J.; Shen, Z.; Zhang, M.; Tang, L.; et al. A rationally designed semiconducting polymer brush for nir-ii imaging-guided light-triggered remote control of CRISPR/Cas9 genome editing. Adv. Mater. 2019, 31, 1–9. [Google Scholar] [CrossRef]
- Chou, S.J.; Yang, P.; Ban, Q.; Yang, Y.P.; Wang, M.L.; Chien, C.S.; Chen, S.J.; Sun, N.; Zhu, Y.; Liu, H.; et al. Dual supramolecular nanoparticle vectors enable CRISPR/Cas9-mediated knockin of retinoschisin 1 gene—A potential nonviral therapeutic solution for X-linked juvenile retinoschisis. Adv. Sci. 2020, 7, 1–11. [Google Scholar] [CrossRef]
- Lao, Y.H.; Li, M.; Gao, M.A.; Shao, D.; Chi, C.W.; Huang, D.; Chakraborty, S.; Ho, T.C.; Jiang, W.; Wang, H.X.; et al. HPV oncogene manipulation using nonvirally delivered CRISPR/Cas9 or natronobacterium gregoryi argonaute. Adv. Sci. 2018, 5, 1–12. [Google Scholar] [CrossRef]
- Kim, S.M.; Yang, Y.; Oh, S.J.; Hong, Y.; Seo, M.; Jang, M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J. Control. Release 2017, 266, 8–16. [Google Scholar] [CrossRef]
- Zhuang, J.; Tan, J.; Wu, C.; Zhang, J.; Liu, T.; Fan, C.; Li, J.; Zhang, Y. Extracellular vesicles engineered with valency-controlled DNA nanostructures deliver CRISPR/Cas9 system for gene therapy. Nucleic Acids Res. 2020, 48, 8870–8882. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Jaffar Ali, D.; Xu, H.; Kumaravel, S.; Si, K.; Li, Y.; Sun, B.; Ma, J.; Xiao, Z. Epithelial cell -derived microvesicles: A safe delivery platform of CRISPR/Cas9 conferring synergistic anti-tumor effect with sorafenib. Exp. Cell Res. 2020, 392, 112040. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zhang, Z.; Zhao, L.; Qin, Y.; Cai, H.; Geng, Z.; Zhu, X.; Zhang, W.; Zhang, Y.; Tan, J.; et al. Tropism-facilitated delivery of CRISPR/Cas9 system with chimeric antigen receptor-extracellular vesicles against B-cell malignancies. J. Control. Release 2020, 326, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Oh, J.; Shim, G.; Cho, B.; Chang, Y.; Kim, S.; Baek, S.; Kim, H.; Shin, J.; Choi, H.; et al. In vivo neuronal gene editing via CRISPR–Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer’s disease. Nat. Neurosci. 2019, 22, 524–528. [Google Scholar] [CrossRef]
- Shen, Y.; Cohen, J.L.; Nicoloro, S.M.; Kelly, M.; Yenilmez, B.; Henriques, F.; Tsagkaraki, E.; Edwards, Y.J.K.; Hu, X.; Friedline, R.H.; et al. CRISPR-delivery particles targeting nuclear receptor–interacting protein 1 (Nrip1) in adipose cells to enhance energy expenditure. J. Biol. Chem. 2018, 293, 17291–17305. [Google Scholar] [CrossRef] [Green Version]
- Montagna, C.; Petris, G.; Casini, A.; Maule, G.; Franceschini, G.M.; Zanella, I.; Conti, L.; Arnoldi, F.; Burrone, O.R.; Zentilin, L.; et al. VSV-G-enveloped vesicles for traceless delivery of CRISPR-Cas9. Mol. Ther. Nucleic Acids 2018, 12, 453–462. [Google Scholar] [CrossRef] [Green Version]
- Mangeot, P.E.; Risson, V.; Fusil, F.; Marnef, A.; Laurent, E.; Blin, J.; Mournetas, V.; Massouridès, E.; Sohier, T.J.M.; Corbin, A.; et al. Genome editing in primary cells and in vivo using viral-derived nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Gee, P.; Lung, M.S.Y.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M.; et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat. Commun. 2020, 11, 4–20. [Google Scholar] [CrossRef] [Green Version]
- Buck, J.; Grossen, P.; Cullis, P.R.; Huwyler, J.; Witzigmann, D. Lipid-based DNA therapeutics: Hallmarks of non-viral gene delivery. ACS Nano 2019, 13, 3754–3782. [Google Scholar] [CrossRef]
- Noh, S.M.; Han, S.E.; Shim, G.; Lee, K.E.; Kim, C.W.; Han, S.S.; Choi, Y.; Kim, Y.K.; Kim, W.K.; Oh, Y.K. Tocopheryl oligochitosan-based self assembling oligomersomes for siRNA delivery. Biomaterials 2011, 32, 849–857. [Google Scholar] [CrossRef]
- Mohammadinejad, R.; Dehshahri, A.; Sagar Madamsetty, V.; Zahmatkeshan, M.; Tavakol, S.; Makvandi, P.; Khorsandi, D.; Pardakhty, A.; Ashrafizadeh, M.; Ghasemipour Afshar, E.; et al. In vivo gene delivery mediated by non-viral vectors for cancer therapy. J. Control. Release 2020, 325, 249–275. [Google Scholar] [CrossRef] [PubMed]
- Möller, K.; Müller, K.; Engelke, H.; Bräuchle, C.; Wagner, E.; Bein, T. Highly efficient siRNA delivery from core-shell mesoporous silica nanoparticles with multifunctional polymer caps. Nanoscale 2016, 8, 4007–4019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseini, E.S.; Nikkhah, M.; Hosseinkhani, S. Cholesterol-rich lipid-mediated nanoparticles boost of transfection efficiency, utilized for gene editing by crispr-cas9. Int. J. Nanomed. 2019, 14, 4353–4366. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Shan, X.; Luo, C.; He, Z. Emerging nanoparticulate drug delivery systems of metformin. J. Pharm. Investig. 2020, 50, 219–230. [Google Scholar] [CrossRef]
- Giugliani, R. Mucopolysacccharidoses: From understanding to treatment, a century of discoveries. Genet. Mol. Biol. 2012, 35, 924–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dharmalingam, P.; Rachamalla, H.K.R.; Lohchania, B.; Bandlamudi, B.; Thangavel, S.; Murugesan, M.K.; Banerjee, R.; Chaudhuri, A.; Voshavar, C.; Marepally, S. Green transfection: Cationic lipid nanocarrier system derivatized from vegetable fat, palmstearin enhances nucleic acid transfections. ACS Omega 2017, 2, 7892–7903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zangi, L.; Lui, K.O.; Von Gise, A.; Ma, Q.; Ebina, W.; Ptaszek, L.M.; Später, D.; Xu, H.; Tabebordbar, M.; Gorbatov, R.; et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013, 31, 898–907. [Google Scholar] [CrossRef] [Green Version]
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics 2020, 12, 102. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Ji, W.; Hall, J.M.; Hu, Q.; Wang, C.; Beisel, C.L.; Gu, Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem. Int. Ed. 2015, 54, 12029–12033. [Google Scholar] [CrossRef]
- Ehlén, Å.; Martin, C.; Miron, S.; Julien, M.; Theillet, F.X.; Ropars, V.; Sessa, G.; Beaurepere, R.; Boucherit, V.; Duchambon, P.; et al. Proper chromosome alignment depends on BRCA2 phosphorylation by PLK1. Nat. Commun. 2020, 11, 1819. [Google Scholar] [CrossRef] [Green Version]
- Ainalem, M.L.; Bartles, A.; Muck, J.; Dias, R.S.; Carnerup, A.M.; Zink, D.; Nylander, T. DNA compaction induced by a cationic polymer or surfactant impact gene expression and DNA degradation. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eoh, J.; Gu, L. Biomaterials as vectors for the delivery of CRISPR-Cas9. Biomater. Sci. 2019, 7, 1240–1261. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wan, T.; Wang, H.; Zhang, S.; Ping, Y.; Cheng, Y. A boronic acid–rich dendrimer with robust and unprecedented efficiency for cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci. Adv. 2019, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, F.; Huang, X.; Gao, X.; Xie, M.; Pan, G.; Li, Q.; Song, J.; Zhu, X.; Zhang, C. A non-cationic nucleic acid nanogel for the delivery of the CRISPR/Cas9 gene editing tool. Nanoscale 2019, 11, 17211–17215. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Sun, W.; Lin, S.; Jin, R.; Ma, L.; Liu, Y. Cytosolic delivery of CRISPR/Cas9 ribonucleoproteins for genome editing using chitosan-coated red fluorescent protein. Chem. Commun. 2019, 55, 4707–4710. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, A.; Kouhbanani, M.A.J.; Beheshtkhoo, N.; Beigi, V.; Mousavi, S.M.; Hashemi, S.A.R.; Karimi Zade, A.; Amani, A.M.; Savardashtaki, A.; Mirzaei, E.; et al. Polyethylenimine-based nanocarriers in co-delivery of drug and gene: A developing horizon. Nano Rev. Exp. 2018, 9, 1488497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, W.J.; Chen, L.C.; Ho, H.O.; Lin, H.L.; Sheu, M.T. Stearyl polyethylenimine complexed with plasmids as the core of human serum albumin nanoparticles noncovalently bound to CRISPR/Cas9 plasmids or siRNA for disrupting or silencing PD-L1 expression for immunotherapy. Int. J. Nanomed. 2018, 13, 7079–7094. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Wan, T.; Chen, Y.; Chen, Y.; Sun, H.; Cao, T.; Songyang, Z.; Tang, G.; Wu, C.; Ping, Y.; et al. Cationic polymer-mediated CRISPR/Cas9 plasmid delivery for genome editing. Macromol. Rapid Commun. 2019, 40, 1–8. [Google Scholar] [CrossRef]
- Cao, A.; Galanello, R. Beta-thalassemia. Genet. Med. 2010, 12, 61–76. [Google Scholar] [CrossRef] [Green Version]
- Müller, K.; Faeh, C.; Diederich, F. Fluorine in pharmaceuticals: Looking beyond intuition. Science 2007, 317, 1881–1886. [Google Scholar] [CrossRef] [Green Version]
- Gillis, E.P.; Eastman, K.J.; Hill, M.D.; Donnelly, D.J.; Meanwell, N.A. Applications of Fluorine in Medicinal Chemistry. J. Med. Chem. 2015, 58, 8315–8359. [Google Scholar] [CrossRef] [PubMed]
- Rong, G.; Wang, C.; Chen, L.; Yan, Y.; Cheng, Y. Fluoroalkylation promotes cytosolic peptide delivery. Sci. Adv. 2020, 6, eaaz1774. [Google Scholar] [CrossRef] [PubMed]
- Percec, V.; Imam, M.R.; Peterca, M.; Leowanawat, P. Self-organizable vesicular columns assembled from polymers dendronized with semifluorinated janus dendrimers act as reverse thermal actuators. J. Am. Chem. Soc. 2012, 134, 4408–4420. [Google Scholar] [CrossRef] [PubMed]
- Papeo, G. MutT Homolog 1 (MTH1): The silencing of a target. J. Med. Chem. 2016, 59, 2343–2345. [Google Scholar] [CrossRef]
- Abedi-Gaballu, F.; Dehghan, G.; Ghaffari, M.; Yekta, R.; Abbaspour-Ravasjani, S.; Baradaran, B.; Dolatabadi, J.E.N.; Hamblin, M.R. PAMAM dendrimers as efficient drug and gene delivery nanosystems for cancer therapy. Appl. Mater. Today 2018, 12, 177–190. [Google Scholar] [CrossRef]
- Li, J.; Cai, C.; Li, J.; Li, J.; Li, J.; Sun, T.; Wang, L.; Wu, H.; Yu, G. Chitosan-based nanomaterials for drug delivery. Molecules 2018, 23, 2661. [Google Scholar] [CrossRef] [Green Version]
- Santos-Carballal, B.; Fernández, E.F.; Goycoolea, F.M. Chitosan in non-viral gene delivery: Role of structure, characterization methods, and insights in cancer and rare diseases therapies. Polymers 2018, 10, 444. [Google Scholar] [CrossRef] [Green Version]
- Peers, S.; Montembault, A.; Ladavière, C. Chitosan hydrogels for sustained drug delivery. J. Control. Release 2020, 326, 150–163. [Google Scholar] [CrossRef]
- Lim, S.M.; Song, D.K.; Oh, S.H.; Lee-Yoon, D.S.; Bae, E.H.; Lee, J.H. In vitro and in vivo degradation behavior of acetylated chitosan porous beads. J. Biomater. Sci. Polym. Ed. 2008, 19, 453–466. [Google Scholar] [CrossRef]
- Elsharkasy, O.M.; Nordin, J.Z.; Hagey, D.W.; de Jong, O.G.; Schiffelers, R.M.; Andaloussi, S.E.L.; Vader, P. Extracellular vesicles as drug delivery systems: Why and how? Adv. Drug Deliv. Rev. 2020. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, X.; Sansanaphongpricha, K.; Myers, I.; Chen, H.; Yuan, H.; Sun, D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017, 38, 754–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeedi, S.; Israel, S.; Nagy, C.; Turecki, G. The emerging role of exosomes in mental disorders. Transl. Psychiatry 2019, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome theranostics: Biology and translational medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wu, J.; Gu, W.; Huang, Y.; Tong, Z.; Huang, L.; Tan, J. Exosome–liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv. Sci. 2018, 5, 1–9. [Google Scholar] [CrossRef]
- Ye, Y.; Zhang, X.; Xie, F.; Xu, B.; Xie, P.; Yang, T.; Shi, Q.; Zhang, C.Y.; Zhang, Y.; Chen, J.; et al. An engineered exosome for delivering sgRNA:Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomater. Sci. 2020, 8, 2966–2976. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Doyle, L.M.; Wang, M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Kanada, M.; Bachmann, M.H.; Hardy, J.W.; Frimannson, D.O.; Bronsart, L.; Wang, A.; Sylvester, M.D.; Schmidt, T.L.; Kaspar, R.L.; Butte, M.J.; et al. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc. Natl. Acad. Sci. USA 2015, 112, E1433–E1442. [Google Scholar] [CrossRef] [Green Version]
- Tesauro, D.; Accardo, A.; Diaferia, C.; Milano, V.; Guillon, J.; Ronga, L.; Rossi, F. Peptide-based drug-delivery systems in biotechnological applications: Recent advances and perspectives. Molecules 2019, 24, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, S.H.; Jang, J.; Cheon, D.H.; Chong, S.-E.; Ahn, J.H.; Hyun, S.; Yu, J.; Lee, Y. pH-Activatable cell penetrating peptide dimers for potent delivery of anticancer drug to triple-negative breast cancer. J. Control. Release 2020. [Google Scholar] [CrossRef] [PubMed]
- Panda, J.J.; Chauhan, V.S. Short peptide based self-assembled nanostructures: Implications in drug delivery and tissue engineering. Polym. Chem. 2014, 5, 4418–4436. [Google Scholar] [CrossRef]
- Miyamoto, T.; Tsuchiya, K.; Numata, K. Dual peptide-based gene delivery system for the efficient transfection of plant callus cells. Biomacromolecules 2020, 21, 2735–2744. [Google Scholar] [CrossRef]
- Qiu, Y.; Man, R.C.H.; Liao, Q.; Kung, K.L.K.; Chow, M.Y.T.; Lam, J.K.W. Effective mRNA pulmonary delivery by dry powder formulation of PEGylated synthetic KL4 peptide. J. Control. Release 2019, 314, 102–115. [Google Scholar] [CrossRef]
- Yin, L.; Yuvienco, C.; Montclare, J.K. Protein based therapeutic delivery agents: Contemporary developments and challenges. Biomaterials 2017, 134, 91–116. [Google Scholar] [CrossRef]
- Hong, S.; Choi, D.W.; Kim, H.N.; Park, C.G.; Lee, W.; Park, H.H. Protein-based nanoparticles as drug delivery systems. Pharmaceutics 2020, 12, 604. [Google Scholar] [CrossRef]
- Yin, J.; Hou, S.; Wang, Q.; Bao, L.; Liu, D.; Yue, Y.; Yao, W.; Gao, X. Microenvironment-responsive delivery of the Cas9 RNA-guided endonuclease for efficient genome editing. Bioconjug. Chem. 2019, 30, 898–906. [Google Scholar] [CrossRef]
- Timin, A.S.; Muslimov, A.R.; Lepik, K.V.; Epifanovskaya, O.S.; Shakirova, A.I.; Mock, U.; Riecken, K.; Okilova, M.V.; Sergeev, V.S.; Afanasyev, B.V.; et al. Efficient gene editing via non-viral delivery of CRISPR–Cas9 system using polymeric and hybrid microcarriers. Nanomedicine 2018, 14, 97–108. [Google Scholar] [CrossRef]
- Zhu, X.; Lv, M.M.; Liu, J.W.; Yu, R.Q.; Jiang, J.H. DNAzyme activated protein-scaffolded CRISPR-Cas9 nanoassembly for genome editing. Chem. Commun. 2019, 55, 6511–6514. [Google Scholar] [CrossRef]
- He, X.Y.; Ren, X.H.; Peng, Y.; Zhang, J.P.; Ai, S.L.; Liu, B.Y.; Xu, C.; Cheng, S.X. Aptamer/peptide-functionalized genome-editing system for effective immune restoration through reversal of pd-l1-mediated cancer immunosuppression. Adv. Mater. 2020, 32, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liebsch, F.; Kulic, L.; Teunissen, C.; Shobo, A.; Ulku, I.; Engelschalt, V.; Hancock, M.A.; van der Flier, W.M.; Kunach, P.; Rosa-Neto, P.; et al. Aβ34 is a BACE1-derived degradation intermediate associated with amyloid clearance and Alzheimer’s disease progression. Nat. Commun. 2019, 10, 2240. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, S.; Kwaku Dad, A.B.; Beloor, J.; Gopalappa, R.; Lee, S.K.; Kim, H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derakhshankhah, H.; Jafari, S. Cell penetrating peptides: A concise review with emphasis on biomedical applications. Biomed. Pharmacother. 2018, 108, 1090–1096. [Google Scholar] [CrossRef]
- Derouchey, J.; Hoover, B.; Rau, D.C. A comparison of DNA compaction by arginine and lysine peptides: A physical basis for arginine rich protamines. Biochemistry 2013, 52, 3000–3009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Nolte, R.J.M.; Cornelissen, J.J.L.M. Virus-based nanocarriers for drug delivery. Adv. Drug Deliv. Rev. 2012, 64, 811–825. [Google Scholar] [CrossRef]
- Lee, E.B.; Kim, J.H.; Hur, W.; Choi, J.E.; Kim, S.M.; Park, D.J.; Kang, B.Y.; Lee, G.W.; Yoon, S.K. Liver-specific gene delivery using engineered virus-like particles of hepatitis e virus. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Nikolic, J.; Belot, L.; Raux, H.; Legrand, P.; Gaudin, Y.; Albertini, A.A. Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, T.H.; de Oliveira Freitas, L.B.; Fernandes, R.S.; dos Santos, V.M.; Resende, J.M.; Cardoso, V.N.; de Barros, A.L.B.; de Sousa, E.M.B. Boron nitride nanotube-CREKA peptide as an effective target system to metastatic breast cancer. J. Pharm. Investig. 2020, 50, 469–480. [Google Scholar] [CrossRef]
- Rosenblum, D.; Joshi, N.; Tao, W.; Karp, J.M.; Peer, D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018, 9, 1410. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wang, L.; Xie, Y.; Wang, P.; Deng, S.; Qin, A.; Zhang, J.; Yu, X.; Zheng, W.; Jiang, X. Triple-targeting delivery of CRISPR/Cas9 to reduce the risk of cardiovascular diseases. Angew. Chem. Int. Ed. 2019, 58, 12404–12408. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Abdeen, A.A.; Wang, Y.; Shahi, P.K.; Robertson, S.; Xie, R.; Suzuki, M.; Pattnaik, B.R.; Saha, K.; Gong, S. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat. Nanotechnol. 2019, 14, 974–980. [Google Scholar] [CrossRef] [PubMed]
- He, Z.Y.; Zhang, Y.G.; Yang, Y.H.; Ma, C.C.; Wang, P.; Du, W.; Li, L.; Xiang, R.; Song, X.R.; Zhao, X.; et al. In vivo ovarian cancer gene therapy using CRISPR-Cas9. Hum. Gene Ther. 2018, 29, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Lo, J.H.; Rananaware, S.; Downing, M.; Panda, A.; Tai, M.; Raghavan, S.; Fleming, H.E.; Bhatia, S.N. Non-viral delivery of CRISPR/Cas9 complex using CRISPR-GPS nanocomplexes. Nanoscale 2019, 11, 21317–21323. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Liu, F.; Chen, Y.; Liu, J.; Wang, X.; Chen, A.T.; Deng, G.; Zhang, H.; Liu, J.; Hong, Z.; et al. Targeted delivery of CRISPR/Cas9-Mediated cancer gene therapy via liposome-templated hydrogel nanoparticles. Adv. Funct. Mater. 2017, 27, 1–9. [Google Scholar] [CrossRef]
- He, X.Y.; Liu, B.Y.; Peng, Y.; Zhuo, R.X.; Cheng, S.X. Multifunctional vector for delivery of genome editing plasmid targeting β-catenin to remodulate cancer cell properties. ACS Appl. Mater. Interfaces 2019, 11, 226–237. [Google Scholar] [CrossRef]
- Liu, B.Y.; He, X.Y.; Zhuo, R.X.; Cheng, S.X. Reversal of tumor malignization and modulation of cell behaviors through genome editing mediated by a multi-functional nanovector. Nanoscale 2018, 10, 21209–21218. [Google Scholar] [CrossRef]
- Liang, C.; Li, F.; Wang, L.; Zhang, Z.K.; Wang, C.; He, B.; Li, J.; Chen, Z.; Shaikh, A.B.; Liu, J.; et al. Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma. Biomaterials 2017, 147, 68–85. [Google Scholar] [CrossRef] [Green Version]
- Mamidyala, S.K.; Dutta, S.; Chrunyk, B.A.; Préville, C.; Wang, H.; Withka, J.M.; McColl, A.; Subashi, T.A.; Hawrylik, S.J.; Griffor, M.C.; et al. Glycomimetic ligands for the human asialoglycoprotein receptor. J. Am. Chem. Soc. 2012, 134, 1978–1981. [Google Scholar] [CrossRef]
- Perrone, F.; Craparo, E.F.; Cemazar, M.; Kamensek, U.; Drago, S.E.; Dapas, B.; Scaggiante, B.; Zanconati, F.; Bonazza, D.; Grassi, M.; et al. Targeted delivery of siRNAs against hepatocellular carcinoma-related genes by a galactosylated polyaspartamide copolymer. J. Control. Release 2020. [Google Scholar] [CrossRef]
- Nair, J.K.; Willoughby, J.L.S.; Chan, A.; Charisse, K.; Alam, M.R.; Wang, Q.; Hoekstra, M.; Kandasamy, P.; Kelin, A.V.; Milstein, S.; et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014, 136, 16958–16961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouet, R.; Thuma, B.A.; Roy, M.D.; Lintner, N.G.; Rubitski, D.M.; Finley, J.E.; Wisniewska, H.M.; Mendonsa, R.; Hirsh, A.; De Oñate, L.; et al. Receptor-mediated delivery of CRISPR-Cas9 endonuclease for cell-type-specific gene editing. J. Am. Chem. Soc. 2018, 140, 6596–6603. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xie, H.; Liu, Y.; Xia, C.; Cun, X.; Long, Y.; Chen, X.; Deng, M.; Guo, R.; Zhang, Z.; et al. Knockdown of hypoxia-inducible factor-1 alpha by tumor targeted delivery of CRISPR/Cas9 system suppressed the metastasis of pancreatic cancer. J. Control. Release 2019, 304, 204–215. [Google Scholar] [CrossRef]
- Jeddi, I.; Saiz, L. Three-dimensional modeling of single stranded DNA hairpins for aptamer-based biosensors. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alshaer, W.; Hillaireau, H.; Fattal, E. Aptamer-guided nanomedicines for anticancer drug delivery. Adv. Drug Deliv. Rev. 2018, 134, 122–137. [Google Scholar] [CrossRef]
- Wang, H.X.; Song, Z.; Lao, Y.H.; Xu, X.; Gong, J.; Cheng, D.; Chakraborty, S.; Park, J.S.; Li, M.; Huang, D.; et al. Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc. Natl. Acad. Sci. USA 2018, 115, 4903–4908. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Zhang, L.; Zheng, W.; Cong, L.; Guo, Z.; Xie, Y.; Wang, L.; Tang, R.; Feng, Q.; Hamada, Y.; et al. Thermo-triggered release of CRISPR-Cas9 system by lipid-encapsulated gold nanoparticles for tumor therapy. Angew. Chem. Int. Ed. 2018, 57, 1491–1496. [Google Scholar] [CrossRef]
- Wang, M.; Zuris, J.A.; Meng, F.; Rees, H.; Sun, S.; Deng, P.; Han, Y.; Gao, X.; Pouli, D.; Wu, Q.; et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. USA 2016, 113, 2868–2873. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Cai, J.; Zheng, Y.; Tan, Y.; Wang, Y.; Zhang, Z.; Zheng, C.; Zhao, Y.; Liu, C.; An, Y.; et al. NanoRNP overcomes tumor heterogeneity in cancer treatment. Nano Lett. 2019, 19, 7662–7672. [Google Scholar] [CrossRef]
- Ma, Y.; Han, X.; Quintana Bustamante, O.; Bessa de Castro, R.; Zhang, K.; Zhang, P.; Li, Y.; Liu, Z.; Liu, X.; Ferrari, M.; et al. Highly efficient genome editing of human hematopoietic stem cells via a nano-silicon-blade delivery approach. Integr. Biol. 2017, 9, 548–554. [Google Scholar] [CrossRef] [Green Version]
- Vad, B.S.; Bertelsen, K.; Johansen, C.H.; Pedersen, J.M.; Skrydstrup, T.; Nielsen, N.C.; Otzen, D.E. Pardaxin permeabilizes vesicles more efficiently by pore formation than by disruption. Biophys. J. 2010, 98, 576–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taharabaru, T.; Yokoyama, R.; Higashi, T.; Mohammed, A.F.A.; Inoue, M.; Maeda, Y.; Niidome, T.; Onodera, R.; Motoyama, K. Genome editing in a wide area of the brain using dendrimer-based ternary polyplexes of Cas9 ribonucleoprotein. ACS Appl. Mater. Interfaces 2020, 12, 21386–21397. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ogura, S.; Otabe, T.; Kamegawa, R.; Sato, M.; Kataoka, K.; Miyata, K. Fine-tuning of hydrophobicity in amphiphilic polyaspartamide derivatives for rapid and transient expression of messenger rna directed toward genome engineering in brain. ACS Cent. Sci. 2019, 5, 1866–1875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, S.; Li, X.; Liu, S.; Chen, J.; Li, M.; Chew, S.Y.; Leong, K.W.; Cheng, D. Codelivery of CRISPR-Cas9 and chlorin e6 for spatially controlled tumor-specific gene editing with synergistic drug effects. Sci. Adv. 2020, 6, 1–15. [Google Scholar] [CrossRef]
- Lyu, Y.; He, S.; Li, J.; Jiang, Y.; Sun, H.; Miao, Y.; Pu, K. A photolabile semiconducting polymer nanotransducer for near-infrared regulation of CRISPR/Cas9 gene editing. Angew. Chem. Int. Ed. 2019, 58, 18197–18201. [Google Scholar] [CrossRef]
- Liu, J.; Chang, J.; Jiang, Y.; Meng, X.; Sun, T.; Mao, L.; Xu, Q.; Wang, M. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 2019, 31, 1–7. [Google Scholar] [CrossRef]
- Chen, G.; Ma, B.; Wang, Y.; Gong, S. A universal GSH-responsive nanoplatform for the delivery of DNA, mRNA, and Cas9/sgRNA ribonucleoprotein. ACS Appl. Mater. Interfaces 2018, 10, 18515–18523. [Google Scholar] [CrossRef]
- Qi, Y.; Song, H.; Xiao, H.; Cheng, G.; Yu, B.; Xu, F.J. Fluorinated acid-labile branched hydroxyl-rich nanosystems for flexible and robust delivery of plasmids. Small 2018, 14, 1–13. [Google Scholar] [CrossRef]
- Sun, D.; Sun, Z.; Jiang, H.; Vaidya, A.M.; Xin, R.; Ayat, N.R.; Schilb, A.L.; Qiao, P.L.; Han, Z.; Naderi, A.; et al. Synthesis and evaluation of ph-sensitive multifunctional lipids for efficient delivery of CRISPR/Cas9 in gene editing. Bioconjug. Chem. 2019, 30, 667–678. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, A.; Matsumoto, D.; Kato, Y.; Honda, Y.; Morikawa, M.; Iwata, F.; Kobayashi, T.; Nakamura, C. Direct delivery of Cas9-sgRNA ribonucleoproteins into cells using a nanoneedle array. Appl. Sci. 2019, 9, 965. [Google Scholar] [CrossRef] [Green Version]
- Hansen-Bruhn, M.; de Ávila, B.E.-F.; Beltrán-Gastélum, M.; Zhao, J.; Ramírez-Herrera, D.E.; Angsantikul, P.; Vesterager Gothelf, K.; Zhang, L.; Wang, J. Active intracellular delivery of a Cas9/sgRNA complex using ultrasound-propelled nanomotors. Angew. Chem. Int. Ed. 2018, 57, 2657–2661. [Google Scholar] [CrossRef] [PubMed]
- Canton, I.; Battaglia, G. Endocytosis at the nanoscale. Chem. Soc. Rev. 2012, 41, 2718–2739. [Google Scholar] [CrossRef] [PubMed]
- Iversen, T.G.; Skotland, T.; Sandvig, K. Endocytosis and intracellular transport of nanoparticles: Present knowledge and need for future studies. Nano Today 2011, 6, 176–185. [Google Scholar] [CrossRef]
- Smith, S.A.; Selby, L.I.; Johnston, A.P.R.; Such, G.K. The endosomal escape of nanoparticles: Toward more efficient cellular delivery. Bioconjug. Chem. 2019, 30, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Yuan, X.; Luo, L.; Lu, Y.; Qin, B.; Zhang, J.; Shi, Y.; Zhu, C.; Yang, J.; Li, X.; et al. Appropriate delivery of the CRISPR/Cas9 system through the nonlysosomal route: Application for therapeutic gene editing. Adv. Sci. 2020, 7, 1–11. [Google Scholar] [CrossRef]
- Le, Q.-V.; Suh, J.; Choi, J.; Park, G.T.; Lee, J.W.; Shim, G.; Oh, Y.-K. In situ nanoadjuvant-assembled tumor vaccine for preventing long-term recurrence. ACS Nano 2019, 13, 7442–7462. [Google Scholar] [CrossRef]
- Jin, H.; Kim, M.G.; Ko, S.B.; Kim, D.H.; Lee, B.J.; Macgregor, R.B.J.; Shim, G.; Oh, Y.K. Stemmed DNA nanostructure for the selective delivery of therapeutics. Nanoscale 2018, 10, 7511–7518. [Google Scholar] [CrossRef]
- Bao, Z.; Liu, X.; Liu, Y.; Liu, H.; Zhao, K. Near-infrared light-responsive inorganic nanomaterials for photothermal therapy. Asian J. Pharm. Sci. 2016, 11, 349–364. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Byun, J.; Park, J.; Lee, Y.; Shim, G.; Oh, Y.-K. Biomimetic polymeric nanoparticle-based photodynamic immunotherapy and protection against tumor rechallenge. Biomater. Sci. 2020, 8, 1106–1116. [Google Scholar] [CrossRef]
- Cheng, R.; Feng, F.; Meng, F.; Deng, C.; Feijen, J.; Zhong, Z. Glutathione-responsive nano-vehicles as a promising platform for targeted intracellular drug and gene delivery. J. Control. Release 2011, 152, 2–12. [Google Scholar] [CrossRef]
- Liu, F.; Ma, F.; Wang, Y.; Hao, L.; Zeng, H.; Jia, C.; Wang, Y.; Liu, P.; Ong, I.M.; Li, B.; et al. PKM2 methylation by CARM1 activates aerobic glycolysis to promote tumorigenesis. Nat. Cell Biol. 2017, 19, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.F.; Whittaker, M.R.; Davis, T.P. Glutathione responsive polymers and their application in drug delivery systems. Polym. Chem. 2017, 8, 97–126. [Google Scholar] [CrossRef]
- Kanamala, M.; Wilson, W.R.; Yang, M.; Palmer, B.D.; Wu, Z. Mechanisms and biomaterials in pH-responsive tumour targeted drug delivery: A review. Biomaterials 2016, 85, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shahi, P.K.; Xie, R.; Zhang, H.; Abdeen, A.A.; Yodsanit, N.; Ma, Z.; Saha, K.; Pattnaik, B.R.; Gong, S. A pH-responsive silica–metal–organic framework hybrid nanoparticle for the delivery of hydrophilic drugs, nucleic acids, and CRISPR-Cas9 genome-editing machineries. J. Control. Release 2020, 324, 194–203. [Google Scholar] [CrossRef]
- Edgcomb, S.P.; Murphy, K.P. Variability in the pKa of histidine side-chains correlates with burial within proteins. Proteins Struct. Funct. Genet. 2002, 49, 1–6. [Google Scholar] [CrossRef]
- Fajrial, A.K.; He, Q.Q.; Wirusanti, N.I.; Slansky, J.E.; Ding, X. A review of emerging physical transfection methods for CRISPR/Cas9-mediated gene editing. Theranostics 2020, 10, 5532–5549. [Google Scholar] [CrossRef]
- Du, S.; Liew, S.S.; Li, L.; Yao, S.Q. Bypassing endocytosis: Direct cytosolic delivery of proteins. J. Am. Chem. Soc. 2018, 140, 15986–15996. [Google Scholar] [CrossRef]
- Sun, T.; Dasgupta, A.; Zhao, Z.; Nurunnabi, M.; Mitragotri, S. Physical triggering strategies for drug delivery. Adv. Drug Deliv. Rev. 2020, 158, 36–62. [Google Scholar] [CrossRef]
- Haapaniemi, E.; Botla, S.; Persson, J.; Schmierer, B.; Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018, 24, 927–930. [Google Scholar] [CrossRef] [Green Version]
- Ihry, R.J.; Worringer, K.A.; Salick, M.R.; Frias, E.; Ho, D.; Theriault, K.; Kommineni, S.; Chen, J.; Sondey, M.; Ye, C.; et al. P53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018, 24, 939–946. [Google Scholar] [CrossRef]
- Suk, J.S.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016, 99, 28–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qie, Y.; Yuan, H.; Von Roemeling, C.A.; Chen, Y.; Liu, X.; Shih, K.D.; Knight, J.A.; Tun, H.W.; Wharen, R.E.; Jiang, W.; et al. Surface modification of nanoparticles enables selective evasion of phagocytic clearance by distinct macrophage phenotypes. Sci. Rep. 2016, 6, 1–11. [Google Scholar]
- Söllner, J.F.; Leparc, G.; Hildebrandt, T.; Klein, H.; Thomas, L.; Stupka, E.; Simon, E. An RNA-Seq atlas of gene expression in mouse and rat normal tissues. Sci. Data 2017, 4, 1–11. [Google Scholar] [CrossRef]
- Li, B.; Qing, T.; Zhu, J.; Wen, Z.; Yu, Y.; Fukumura, R.; Zheng, Y.; Gondo, Y.; Shi, L. A comprehensive mouse transcriptomic bodymap across 17 tissues by RNA-seq. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Charlesworth, C.T.; Deshpande, P.S.; Dever, D.P.; Camarena, J.; Lemgart, V.T.; Cromer, M.K.; Vakulskas, C.A.; Collingwood, M.A.; Zhang, L.; Bode, N.M.; et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019, 25, 249–254. [Google Scholar] [CrossRef]
- Riteau, N.; Sher, A. Chitosan: An adjuvant with an unanticipated STING. Immunity 2016, 44, 522–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pizzuto, M.; Bigey, P.; Lachagès, A.M.; Hoffmann, C.; Ruysschaert, J.M.; Escriou, V.; Lonez, C. Cationic lipids as one-component vaccine adjuvants: A promising alternative to alum. J. Control. Release 2018, 287, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Le, Q.V.; Suh, J.; Oh, Y.K. Nanomaterial-based modulation of tumor microenvironments for enhancing chemo/immunotherapy. AAPS J. 2019, 21, 64. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, R.A.; LaMothe, R.A.; Ferrari, J.D.; Zhang, A.H.; Rossi, R.J.; Kolte, P.N.; Griset, A.P.; O’Neil, C.; Altreuter, D.H.; Browning, E.; et al. Polymeric synthetic nanoparticles for the induction of antigen-specific immunological tolerance. Proc. Natl. Acad. Sci. USA 2015, 112, E156–E165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Moon, J.H.; Jeong, S.U.; Jung, H.H.; Park, C.S.; Hwang, B.Y.; Lee, C.K. Induction of antigen-specific immune tolerance using biodegradable nanoparticles containing antigen and dexamethasone. Int. J. Nanomed. 2019, 14, 5229–5242. [Google Scholar] [CrossRef] [Green Version]
- Wu, R.; Pai, A.; Liu, L.; Xing, S.; Lu, Y. NanoTPOT: Enhanced sample preparation for quantitative nanoproteomic analysis. Anal. Chem. 2020, 92, 6235–6240. [Google Scholar] [CrossRef] [PubMed]
- Hamouda, H.; Kaup, M.; Ullah, M.; Berger, M.; Sandig, V.; Tauber, R.; Blanchard, V. Rapid analysis of cell surface N-glycosylation from living cells using mass spectrometry. J. Proteome Res. 2014, 13, 6144–6151. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Oh, D.H.; Oh, Y.K.; Yong, C.S.; Choi, H.G. Effects of solid carriers on the crystalline properties, dissolution and bioavailability of flurbiprofen in solid self-nanoemulsifying drug delivery system (solid SNEDDS). Eur. J. Pharm. Biopharm. 2012, 80, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Shim, G.; Kim, D.; Park, G.T.; Jin, H.; Suh, S.-K.; Oh, Y.-K. Therapeutic gene editing: Delivery and regulatory perspectives. Acta Pharmacol. Sin. 2017, 38, 738–753. [Google Scholar] [CrossRef] [Green Version]







| Material | Composition | Cas9 | sgRNA | Target Gene | Editing Type | Target Disease | Ref |
|---|---|---|---|---|---|---|---|
| Lipid | DOTAP, DOPE, DSPE-PEG, Chol | Plasmid DNA | Plasmid DNA | HPV16E6, E7 | Knockout | Cervical cancer | [30] |
| DOTAP, DOPE, DSPE-PEG | Plasmid DNA | Plasmid DNA | Iduronidase | Knock-in | Mucopolysaccharidosis type I | [31] | |
| cKK-E12, DOPE, Chol, C14-PEG | mRNA | sgRNA | PCSK9 | Knockout | Hypercholesterolemia | [32] | |
| Protamine, chondroitin sulfate, DOTAP, DOPE, DSPE-PEG | Plasmid DNA | Plasmid DNA | PLK1 | Knockout | Melanoma | [33] | |
| Gold nanoparticle, TAT, DOTAP, DOPE, DSPE-PEG | Protein | Plasmid DNA | PLK1 | Knockout | Melanoma | [34] | |
| MSN, DOTAP, DOPE, DSPE-PEG, Chol | Protein | sgRNA | PCSK9, Apolipo-protein C3, Angiopoietin-like 3 | Knockout | Hypercholesterolemia | [35] | |
| Polymer | PEGylated hyaluronan polymer with R8-RGD tandem peptide, fluorinated polyethyleneimine | Plasmid DNA | Plasmid DNA | MTH1 | Knockout | Ovarian cancer | [36] |
| Semiconducting polymers composed of eicosane, PEG, diketopyrrolopyrrole and fluorinated PEI | Plasmid DNA | Plasmid DNA | MTH1 | Knockout | - | [37] | |
| Branched β-CD-PEI/Ad-PAMAM/Ad-PEG-TAT | Plasmid DNA | Plasmid DNA | Retinoschisin 1, GFP | Knock-in | X-linked juvenile retinoschisis | [38] | |
| Quaternary ammonium-terminated poly(propylene oxide)/Pluronic F127 | Plasmid DNA | Plasmid DNA | HPV18-E7 | Knockout | - | [39] | |
| Extracellular vesicle | Exosome from SKOV3 | Protein | sgRNA | Poly (ADP-ribose) polymerase 1 | Knockout | Ovarian cancer | [40] |
| Exosome modified with TLS11a aptamer-Chol | Protein | sgRNA | WNT10B | Knockout | Hepatocellular cancer | [41] | |
| Microvesicle | Protein | sgRNA | IQGAP1 | Knockout | Hepatocellular cancer | [42] | |
| Microvesicle with anti-CD19 CAR | Protein | sgRNA | MYC | Knockout | Lymphoma | [43] | |
| Peptide/protein | R7L10 peptide-based micelle | Protein | sgRNA | β-secretase 1 | Knockout | Alzheimer’s disease | [44] |
| Poly-L-arginine | Protein | sgRNA | GFP, Nuclear Receptor Interacting Protein 1 | Knockout | Obesity | [45] | |
| VLP composed of VSV-G | Protein | sgRNA | GFP | Knockout | - | [46] | |
| VLP composed of Gag-Cas9 fusion, VSV-G, BaEVRLess, sgRNA | Protein | sgRNA | EMX1, DDX3, Tyrosinase, HPD | Knockout | Hereditary tyrosinemia type I (HT1) | [47] | |
| VLP composed of VSV-G, FKBP12-Gag, FRB-Cas9 | Protein | sgRNA | Dystrophin | Knockout | Duchenne muscular dystrophy | [48] |
| Targeting | Material | Composition | Cas9 Type | sgRNA Type | Target Gene | Editing Type | Target Disease | Ref |
|---|---|---|---|---|---|---|---|---|
| Chemical ligand-based targeting | Lipid | Lipid shell composed of Gal-PEG-DSPE, DOTAP, DOPE, Chol, TAT with AuNP | Protein | sgRNA | PCSK9 | Knockout | High LDL cholesterol | [111] |
| Polymer | Polymeric nanoparticle composed of cationic, anionic, imidazole, biodegradable crosslinker, acrylate-mPEG-ATRA | Protein | sgRNA | Stop-tdTomato | Knockout | - | [112] | |
| Lipid | Liposome composed of DOTAP, cholesterol, folate-PEG-succinyl-Chol | Plasmid DNA | Plasmid DNA | DNMT1 | Knockout | Ovarian cancer | [113] | |
| Peptide-based targeting | Lipid | Liposome-templated hydrogel nanoparticle composed of palmitoyl-transportan-iRGD | Protein | sgRNA | GFP | Knockout | - | [114] |
| Lipid | Liposome-templated hydrogel nanoparticle composed of PEI-CD, PEI-AD, mHph3, iRGD | Protein | sgRNA | PLK-1 | Knockout | Brain cancer | [115] | |
| Polymer | Core-shell polymeric nanoparticle composed of PF33, hyaluronan, RGD-R8-PEG | Plasmid DNA | Plasmid DNA | MTH1 | Knockout | Ovarian cancer | [36] | |
| Antibody and aptamer ligand-based targeting | Protein | Protamine-based nanoparticle composed of CaCO3, As1411/TAT functionalized carboxymethyl chitosan | Plasmid DNA | Plasmid DNA | CTNNB1 | Knockout | - | [116] |
| Protein | Protamine-based nanoparticle composed of CaCO3, As1411/mucin 1 aptamer functionalized heparin | Plasmid DNA | Plasmid DNA | Focal adhesion kinase | Knockout | - | [117] | |
| Polymer | Peptide-lipid micelle composed of PEG-PEI-Chol, LC09 aptamer | Plasmid DNA | Plasmid DNA | VEGF-A | Knockout | Osteo-sarcoma | [118] |
| Strategy | Mechanism | Material | Composition | Cas9 Type | sgRNA Type | Target Gene | Editing Type | Ref |
|---|---|---|---|---|---|---|---|---|
| Endosomal escape | Cationic charge-induced membrane disruption | Liposome | Pardaxin peptide-modified cationic liposome | Plasmid DNA | Plasmid DNA | CDC6 | Knockout | [131] |
| Polymer | Glucuronylglucosyl-β-cyclodextrin conjugated dendrimer | Protein | sgRNA | Rosa26 | Knockout | [132] | ||
| Polymer | Cationic diethylenetriamine-conjugated amphiphilic polyaspartamide derivatives | mRNA | sgRNA | Luciferase | Knockout | [133] | ||
| Polypeptide/polymer | α-helical polypeptide incorporating PEG-polythymine 40-based nanocomplex | Plasmid DNA | Plasmid DNA | PLK1 | Knockout | [126] | ||
| Photo-responsive delivery | Photodynamic effect | Polymer | Chlorin e6-loaded/cationic iRGD-modified copolymer-coated polymeric micelle | Protein | sgRNA | Nrf2 | Knockout | [134] |
| Polymer | PEI-thioester-semiconducting polymer | Plasmid DNA | Plasmid DNA | GFP | Knockout | [135] | ||
| Photothermal effect | Lipid | Lipid coated-gold NP-TAT peptide | Plasmid DNA | Plasmid DNA | PLK-1 | Knockout | [127] | |
| Polymer | Semiconducting polymers composed of eicosane, PEG, diketopyrrolopyrrole, and fluorinated PEI | Plasmid DNA | Plasmid DNA | MTH1 | Knockout | [37] | ||
| GSH-responsive delivery | GSH-mediated bond cleavage | Lipid | Cationic lipid nanoparticle composed of 8-O14B, DOPE, Chol, C16-PEG-ceramide | Protein | sgRNA | GFP | Knockout | [128] |
| Lipid | Cationic lipid nanoparticle composed of BAMEA-O16B, DOPE, DSPE-PEG, Chol | mRNA | sgRNA | PCSK9 | Knockout | [136] | ||
| Polymer | Cationic block copolymer composed of poly(Asp-AED-ICA)-PEG | Protein | sgRNA | mCherry | Knockout | [137] | ||
| Polymer | Polymeric nanoparticle composed of cationic, anionic, imidazole, biodegradable crosslinker, acrylate-mPEG-ATRA | Protein | sgRNA | Stop-tdTomato | Knockout | [112] | ||
| pH-responsive delivery | pH-dependent degradation | Polymer | Ortho ester polymer/PEI | Protein | sgRNA | Survivin | Knockout | [138] |
| Polymer | PLys100-CAmPEG77 | Protein | sgRNA | STAT3, RUNX1 | Knockout | [129] | ||
| pH-dependent charge reversion | Lipid | 1-Aminoethyl)iminobis [N-(oleoylcysteinyl-1- aminoethyl)propionamide lipid | Plasmid DNA | Plasmid DNA | GFP | Knockout | [139] | |
| Physical delivery | Mechanical membrane deformation | Nanoneedle | Silicon based atomic force microscope cantilever type nanoneedle | Protein | sgRNA | GFP, Nestin | Knockout | [140] |
| Nanoblade | Photolithography and reactive ion etching fabricated microfluidic chip with silicon nanoblade | Protein | sgRNA | C/EBPα | Knockout | [130] | ||
| Ultrasound mediated membrane penetration | Metal | Cysteine modified asymmetric gold nanowire | Protein | sgRNA | GFP | Knockout | [141] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.; Le, Q.-V.; Wu, Y.; Park, J.; Oh, Y.-K. Nanovesicle-Mediated Delivery Systems for CRISPR/Cas Genome Editing. Pharmaceutics 2020, 12, 1233. https://doi.org/10.3390/pharmaceutics12121233
Kim D, Le Q-V, Wu Y, Park J, Oh Y-K. Nanovesicle-Mediated Delivery Systems for CRISPR/Cas Genome Editing. Pharmaceutics. 2020; 12(12):1233. https://doi.org/10.3390/pharmaceutics12121233
Chicago/Turabian StyleKim, Dongyoon, Quoc-Viet Le, Yina Wu, Jinwon Park, and Yu-Kyoung Oh. 2020. "Nanovesicle-Mediated Delivery Systems for CRISPR/Cas Genome Editing" Pharmaceutics 12, no. 12: 1233. https://doi.org/10.3390/pharmaceutics12121233