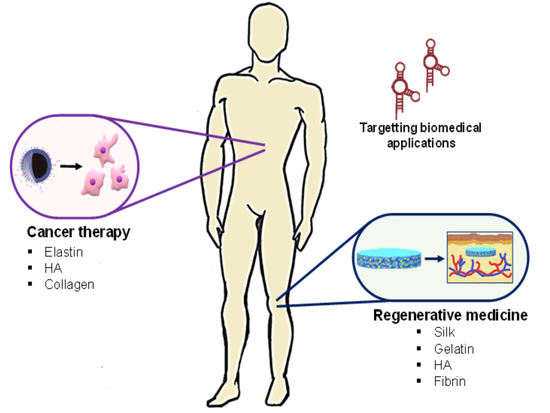
Aptamer-Functionalized Natural Protein-Based Polymers as Innovative Biomaterials
Abstract
:1. Introduction
2. Biomaterials
2.1. Collagen and Gelatin
2.1.1. Cancer Therapy
2.1.2. Wound Healing
2.1.3. Biosensing Application: Current Advances and Progress of Using Natural Protein-Based Polymers
2.2. Elastin
2.2.1. Cancer Therapy
2.2.2. Other Applications
2.3. Fibrinogen and Fibrin
2.3.1. Tissue Regeneration
2.3.2. Cancer Therapy
2.4. Silk
Biosensing Application: Current Advances and Progress of Using Natural Protein-Based Polymers
2.5. Hyaluronic Acid
2.5.1. Glioma
2.5.2. Axons Regeneration
2.6. Hyaluronic Acid Mixtures
2.6.1. Cartilage Repair
2.6.2. Cancer Therapy
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ghosh, M.; Halperin-Sternfeld, M.; Adler-Abramovich, L. Bio Mimicking of Extracellular Matrix. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1174, pp. 371–399. [Google Scholar]
- Ratner, B.D. Biomaterials: Been There, Done That, and Evolving into the Future. Annu. Rev. Biomed. Eng. 2019, 21, 171–191. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.; Hakala, T.A.; Schnaider, L.; Bernardes, G.J.L.; Gazit, E.; Knowles, T.P.J. Biomimetic peptide self-assembly for functional materials. Nat. Rev. Chem. 2020, 4, 615–634. [Google Scholar] [CrossRef]
- Agrahari, V.; Agrahari, V.; Chou, M.L.; Chew, C.H.; Noll, J.; Burnouf, T. Intelligent micro-/nanorobots as drug and cell carrier devices for biomedical therapeutic advancement: Promising development opportunities and translational challenges. Biomaterials 2020, 260. [Google Scholar] [CrossRef]
- Aguado, B.A.; Grim, J.C.; Rosales, A.M.; Watson-Capps, J.J.; Anseth, K.S. Engineering precision biomaterials for personalized medicine. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Sullivan, M.O.; Kiick, K.L. Targeted Drug Delivery via the Use of ECM-Mimetic Materials. Front. Bioeng. Biotechnol. 2020, 8, 69. [Google Scholar] [CrossRef] [Green Version]
- Desai, M.S.; Lee, S.W. Protein-based functional nanomaterial design for bioengineering applications. Wiley Interdiscip. Rev. Nanomedicine Nanobiotechnology 2015, 7, 69–97. [Google Scholar] [CrossRef]
- Wang, Y.; Katyal, P.; Montclare, J.K. Protein-Engineered Functional Materials. Adv. Healthc. Mater. 2019, 8, 1801374. [Google Scholar] [CrossRef]
- Keating, K.W.; Young, E.M. Synthetic biology for bio-derived structural materials. Curr. Opin. Chem. Eng. 2019, 24, 107–114. [Google Scholar] [CrossRef]
- Girotti, A.; Orbanic, D.; Ibáñez-Fonseca, A.; Gonzalez-Obeso, C.; Rodríguez-Cabello, J.C. Recombinant Technology in the Development of Materials and Systems for Soft-Tissue Repair. Adv. Healthc. Mater. 2015, 4, 2423–2455. [Google Scholar] [CrossRef]
- Varanko, A.; Saha, S.; Chilkoti, A. Recent trends in protein and peptide-based biomaterials for advanced drug delivery. Adv. Drug Deliv. Rev. 2020, S0169-409X. [Google Scholar] [CrossRef]
- Costa, F.; Silva, R.; Boccaccini, A.R. Fibrous protein-based biomaterials (silk, keratin, elastin, and resilin proteins) for tissue regeneration and repair. In Peptides and Proteins as Biomaterials for Tissue Regeneration and Repair; Elsevier: Amsterdam, The Netherlands, 2018; pp. 175–204. ISBN 9780081008522. [Google Scholar]
- Chen, F.-M.; Liu, X. Advancing biomaterials of human origin for tissue engineering. Prog. Polym. Sci. 2016, 53, 86–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abascal, N.C.; Regan, L. The past, present and future of protein-based materials. Open Biol. 2018, 8, 180113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katyal, P.; Meleties, M.; Montclare, J.K. Self-Assembled Protein-and Peptide-Based Nanomaterials. ACS Biomater. Sci. Eng. 2019, 5, 4132–4147. [Google Scholar] [CrossRef]
- Sengupta, D.; Heilshorn, S.C. Protein-Engineered Biomaterials: Highly Tunable Tissue Engineering Scaffolds. Tissue Eng. Part B Rev. 2010, 16, 285–293. [Google Scholar] [CrossRef] [Green Version]
- Oliva, N.; Unterman, S.; Zhang, Y.; Conde, J.; Song, H.S.; Artzi, N. Personalized Medicine: Personalizing Biomaterials for Precision Nanomedicine Considering the Local Tissue Microenvironment (Adv. Healthcare Mater. 11/2015). Adv. Healthc. Mater. 2015, 4, 1584–1599. [Google Scholar] [CrossRef]
- Hamley, I.W. Small Bioactive Peptides for Biomaterials Design and Therapeutics. Chem. Rev. 2017, 117, 14015–14041. [Google Scholar] [CrossRef] [Green Version]
- Ho, D.; Quake, S.R.; McCabe, E.R.B.; Chng, W.J.; Chow, E.K.; Ding, X.; Gelb, B.D.; Ginsburg, G.S.; Hassenstab, J.; Ho, C.-M.; et al. Enabling Technologies for Personalized and Precision Medicine. Trends Biotechnol. 2020, 38, 497–518. [Google Scholar] [CrossRef]
- Hao, Z.; Fan, W.; Hao, J.; Wu, X.; Zeng, G.Q.; Zhang, L.J.; Nie, S.F.; Wang, X.D. Efficient delivery of micro RNA to bone-metastatic prostate tumors by using aptamer-conjugated atelocollagen in vitro and in vivo. Drug Deliv. 2016, 23, 874–881. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, J.; Tao, S.J.; Ye, T.; Kong, X.; Ren, L. In Vivo Bio-distribution and Efficient Tumor Targeting of Gelatin/Silica Nanoparticles for Gene Delivery. Nanoscale Res. Lett. 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Xi, Y.; Zhang, Y.; Wu, Q.; Meng, R.; Zheng, B.; Rei, L. Redox-Sensitive Gelatin/Silica-Aptamer Nanogels for Targeted siRNA Delivery. Nanoscale Res. Lett. 2019, 14. [Google Scholar] [CrossRef] [Green Version]
- Soontornworajit, B.; Zhou, J.; Zhang, Z.; Wang, Y. Aptamer-functionalized in situ injectable hydrogel for controlled protein release. Biomacromolecules 2010, 11, 2724–2730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Battig, M.R.; Chen, N.; Gaddes, E.R.; Duncan, K.L.; Wang, Y. Chimeric Aptamer-Gelatin Hydrogels as an Extracellular Matrix Mimic for Loading Cells and Growth Factors. Biomacromolecules 2016, 17, 778–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coyne, J.; Zhao, N.; Olubode, A.; Menon, M.; Wang, Y. Development of hydrogel-like biomaterials via nanoparticle assembly and solid-hydrogel transformation. J. Control. Release 2020, 318, 185–196. [Google Scholar] [CrossRef]
- Derkus, B.; Arslan, Y.E.; Bayrac, A.T.; Kantarcioglu, I.; Emregul, K.C.; Emregul, E. Development of a novel aptasensor using jellyfish collagen as matrix and thrombin detection in blood samples obtained from patients with various neurodisease. Sens. Actuators B Chem. 2016, 228, 725–736. [Google Scholar] [CrossRef]
- Derkus, B.; Arslan, Y.E.; Emregul, K.C.; Emregul, E. Enhancement of aptamer immobilization using egg shell-derived nano-sized spherical hydroxyapatite for thrombin detection in neuroclinic. Talanta 2016, 158, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Zhong, H.; Wang, L.; Liu, Y.; Xu, Y.; Zhang, J.; Xu, C.; He, L.; Wang, H. Facile preparation of a collagen-graphene oxide composite: A sensitive and robust electrochemical aptasensor for determining dopamine in biological samples. Int. J. Biol. Macromol. 2019, 135, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Hamidi-Asl, E.; Dardenne, F.; Blust, R.; De Wael, K. An improved electrochemical aptasensor for chloramphenicol detection based on aptamer incorporated gelatine. Sensors (Switzerland) 2015, 15, 7605–7618. [Google Scholar] [CrossRef] [Green Version]
- Piña, M.J.; Girotti, A.; Santos, M.; Rodríguez-Cabello, J.C.; Arias, F.J. Biocompatible ELR-Based Polyplexes Coated with MUC1 Specific Aptamers and Targeted for Breast Cancer Gene Therapy. Mol. Pharm. 2016, 13, 795–808. [Google Scholar] [CrossRef] [Green Version]
- Piña, M.J.; Girotti, A.; Serrano, S.; Muñoz, R.; Rodríguez-Cabello, J.C.; Arias, F.J. A double safety lock tumor-specific device for suicide gene therapy in breast cancer. Cancer Lett. 2020, 470, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Mie, M.; Matsumoto, R.; Mashimo, Y.; Cass, A.E.G.; Kobatake, E. Development of drug-loaded protein nanoparticles displaying enzymatically-conjugated DNA aptamers for cancer cell targeting. Mol. Biol. Rep. 2019, 46, 261–269. [Google Scholar] [CrossRef]
- Guo, W.; Mashimo, Y.; Kobatake, E.; Mie, M. Construction of DNA-displaying nanoparticles by enzymatic conjugation of DNA and elastin-like polypeptides using a replication initiation protein. Nanotechnology 2020, 31. [Google Scholar] [CrossRef] [PubMed]
- Vogele, K.; Frank, T.; Gasser, L.; Goetzfried, M.A.; Hackl, M.W.; Sieber, S.A.; Simmel, F.C.; Pirzer, T. Towards synthetic cells using peptide-based reaction compartments. Nat. Commun. 2018, 9, 3862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank, T.; Vogele, K.; Dupin, A.; Simmel, F.C.; Pirzer, T. Growth of giant peptide vesicles driven by compartmentalized transcription-translation activity. Chem. A Eur. J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Humenik, M.; Preiß, T.; Gödrich, S.; Papastavrou, G.; Scheibel, T. Functionalized DNA-spider silk nanohydrogels for controlled protein binding and release. Mater. Today Bio 2020, 6, 100045. [Google Scholar] [CrossRef]
- Zhao, N.; Coyne, J.; Xu, M.; Zhang, X.; Suzuki, A.; Shi, P.; Lai, J.; Fong, G.H.; Xiong, N.; Wang, Y. Assembly of Bifunctional Aptamer-Fibrinogen Macromer for VEGF Delivery and Skin Wound Healing. Chem. Mater. 2019, 31, 1006–1015. [Google Scholar] [CrossRef]
- Zhao, N.; Suzuki, A.; Zhang, X.; Shi, P.; Abune, L.; Coyne, J.; Jia, H.; Xiong, N.; Zhang, G.; Wang, Y. Dual Aptamer-Functionalized in Situ Injectable Fibrin Hydrogel for Promotion of Angiogenesis via Codelivery of Vascular Endothelial Growth Factor and Platelet-Derived Growth Factor-BB. ACS Appl. Mater. Interfaces 2019, 11, 18123–18132. [Google Scholar] [CrossRef]
- Juhl, O.; Zhao, N.; Merife, A.B.; Cohen, D.; Friedman, M.; Zhang, Y.; Schwartz, Z.; Wang, Y.; Donahue, H. Aptamer-Functionalized Fibrin Hydrogel Improves Vascular Endothelial Growth Factor Release Kinetics and Enhances Angiogenesis and Osteogenesis in Critically Sized Cranial Defects. ACS Biomater. Sci. Eng. 2019, 5, 6152–6160. [Google Scholar] [CrossRef]
- Zhao, N.; Coyne, J.; Abune, L.; Shi, P.; Lian, X.L.; Zhang, G.; Wang, Y. Exogenous Signaling Molecules Released from Aptamer-Functionalized Hydrogels Promote the Survival of Mesenchymal Stem Cell Spheroids. ACS Appl. Mater. Interfaces 2020, 12, 24599–24610. [Google Scholar] [CrossRef]
- Fujita, H.; Inoue, Y.; Kuwahara, M. Selective incorporation of foreign functionality into fibrin gels through a chemically modified DNA aptamer. Bioorg. Med. Chem. Lett. 2018, 28, 35–39. [Google Scholar] [CrossRef]
- Kuwahara, M.; Fujita, H.; Kataoka, Y.; Nakajima, Y.; Yamada, M.; Sugimoto, N. In situ condensation of an anti-cancer drug into fibrin gel enabling effective inhibition of tumor cell growth. Chem. Commun. 2019, 55, 11679–11682. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, Z.; Zhang, G.; Lin, F.; Liu, Y.; Zhang, Y.; Feng, J.; Chen, W.; Meng, Q.; Chen, L. AS1411 Aptamer/Hyaluronic Acid-Bifunctionalized Microemulsion Co-Loading Shikonin and Docetaxel for Enhanced Antiglioma Therapy. J. Pharm. Sci. 2019, 108, 3684–3694. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.K.; Allen, P.; Song, Y.H.; Wachs, R.A.; Du, Y.; Ellington, A.D.; Schmidt, C.E. Oligonucleotide-functionalized hydrogels for sustained release of small molecule (aptamer) therapeutics. Acta Biomater. 2020, 102, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Song, X.; Li, T.; Chen, J.; Cheng, G.; Yang, L.; Chen, C. Aptamer-Functionalized Bioscaffold Enhances Cartilage Repair by Improving Stem Cell Recruitment in Osteochondral Defects of Rabbit Knees. Am. J. Sports Med. 2019, 47, 2316–2326. [Google Scholar] [CrossRef] [PubMed]
- Galli, C.; Parisi, L.; Piergianni, M.; Smerieri, A.; Passeri, G.; Guizzardi, S.; Costa, F.; Lumetti, S.; Manfredi, E.; Macaluso, G.M. Improved scaffold biocompatibility through anti-Fibronectin aptamer functionalization. Acta Biomater. 2016, 42, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, Z.; Dinarvand, R.; Mottaghitalab, F.; Esfandyari-Manesh, M.; Sayari, E.; Atyabi, F. Aptamer decorated hyaluronan/chitosan nanoparticles for targeted delivery of 5-fluorouracil to MUC1 overexpressing adenocarcinomas. Carbohydr. Polym. 2015, 121, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Varnamkhasti, B.S.; Hosseinzadeh, H.; Azhdarzadeh, M.; Vafaei, S.Y.; Esfandyari-Manesh, M.; Mirzaie, Z.H.; Amini, M.; Ostad, S.N.; Atyabi, F.; Dinarvand, R. Protein corona hampers targeting potential of MUC1 aptamer functionalized SN-38 core-shell nanoparticles. Int. J. Pharm. 2015, 494, 430–444. [Google Scholar] [CrossRef]
- Parenteau-Bareil, R.; Gauvin, R.; Berthod, F. Collagen-based biomaterials for tissue engineering applications. Materials (Basel) 2010, 3, 1863–1887. [Google Scholar] [CrossRef] [Green Version]
- Purcel, G.; Meliţă, D.; Andronescu, E.; Grumezescu, A.M. Collagen-based nanobiomaterials. In Nanobiomaterials in Soft Tissue Engineering; Elsevier: Amsterdam, The Netherlands, 2016; pp. 173–200. ISBN 9780323428651. [Google Scholar]
- Liu, X.; Zheng, C.; Luo, X.; Wang, X.; Jiang, H. Recent advances of collagen-based biomaterials: Multi-hierarchical structure, modification and biomedical applications. Mater. Sci. Eng. C 2019, 99, 1509–1522. [Google Scholar] [CrossRef]
- Olsen, D.; Yang, C.; Bodo, M.; Chang, R.; Leigh, S.; Baez, J.; Carmichael, D.; Perälä, M.; Hämäläinen, E.R.; Jarvinen, M.; et al. Recombinant collagen and gelatin for drug delivery. Adv. Drug Deliv. Rev. 2003, 55, 1547–1567. [Google Scholar] [CrossRef]
- Miranda-Nieves, D.; Chaikof, E.L. Collagen and Elastin Biomaterials for the Fabrication of Engineered Living Tissues. ACS Biomater. Sci. Eng. 2017, 3, 694–711. [Google Scholar] [CrossRef]
- Kawecki, M.; Łabuś, W.; Klama-Baryla, A.; Kitala, D.; Kraut, M.; Glik, J.; Misiuga, M.; Nowak, M.; Bielecki, T.; Kasperczyk, A. A review of decellurization methods caused by an urgent need for quality control of cell-free extracellular matrix’ scaffolds and their role in regenerative medicine. J. Biomed. Mater. Res. Part B Appl. Biomater. 2018, 106, 909–923. [Google Scholar] [CrossRef] [PubMed]
- Lynn, A.K.; Yannas, I.V.; Bonfield, W. Antigenicity and immunogenicity of collagen. J. Biomed. Mater. Res. Part B Appl. Biomater. 2004, 71, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.; Kirk, S.; Tronci, G.; Yang, X.; Wood, D. Influence of telopeptides on the structural and physical properties of polymeric and monomeric acid-soluble type I collagen. Mater. Sci. Eng. C 2017, 77, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Hanai, K.; Kojima, T.; Ota, M.; Onodera, J.; Sawada, N. Effects of Atelocollagen Formulation Containing Oligonucleotide on Endothelial Permeability. J. Drug Deliv. 2012, 2012, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabata, Y.; Ikada, Y. Protein release from gelatin matrices. Adv. Drug Deliv. Rev. 1998, 31, 287–301. [Google Scholar] [CrossRef]
- Dash, R.; Foston, M.; Ragauskas, A.J. Improving the mechanical and thermal properties of gelatin hydrogels cross-linked by cellulose nanowhiskers. Carbohydr. Polym. 2013, 91, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Wei, K.; Lin, S.; Xu, Z.; Sun, Y.; Shi, P.; Li, G.; Bian, L. Mechanically resilient, injectable, and bioadhesive supramolecular gelatin hydrogels crosslinked by weak host-guest interactions assist cell infiltration and in situ tissue regeneration. Biomaterials 2016, 101, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Hannon, G.J.; Rossi, J.J. Unlocking the potential of the human genome with RNA interference. Nature 2004, 431, 371–378. [Google Scholar] [CrossRef]
- Shim, G.; Kim, M.-G.; Park, J.Y.; Oh, Y.-K. Small interfering RNAs (siRNAs) as cancer therapeutics. In Biomaterials for Cancer Therapeutics; Elsevier: Amsterdam, The Netherlands, 2013; pp. 237–269. ISBN 9780857096647. [Google Scholar]
- Ochiya, T.; Nagahara, S.; Sano, A.; Itoh, H.; Terada, M. Biomaterials for Gene Delivery Atelocollagen-mediated Controlled Release of Molecular Medicines. Curr. Gene Ther. 2006, 1, 31–52. [Google Scholar] [CrossRef]
- Mundy, G.R. Metastasis to bone: Causes, consequences and therapeutic opportunities. Nat. Rev. Cancer 2002, 2, 584–593. [Google Scholar] [CrossRef]
- Teo, M.Y.; Morris, M.J. Prostate-Specific Membrane Antigen–Directed Therapy for Metastatic Castration-Resistant Prostate Cancer. Cancer J. 2016, 22, 347–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Rajasekaran, A.K.; Moy, P.; Xia, Y.; Kim, S.; Navarro, V.; Rahmati, R.; Bander, N.H. Constitutive and antibody-induced internalization of prostate-specific membrane antigen. Cancer Res. 1998, 58, 4055–4060. [Google Scholar] [PubMed]
- Aqeilan, R.I.; Calin, G.A.; Croce, C.M. MiR-15a and miR-16-1 in cancer: Discovery, function and future perspectives. Cell Death Differ. 2010, 17, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.Y.; Zhao, Y.; Ren, L.; Jin, L.H.; Sun, L.P.; Yin, P.; Zhang, Y.F.; Zhang, Q.Q. Novel gelatin-siloxane nanoparticles decorated by Tat peptide as vectors for gene therapy. Nanotechnology 2008, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Y.; Wang, Z.Y.; Wen, F.; Ren, L.; Li, J.; Teoh, S.H.; Thian, E.S. Gelatin-siloxane nanoparticles to deliver nitric oxide for vascular cell regulation: Synthesis, cytocompatibility, and cellular responses. J. Biomed. Mater. Res. Part A 2015, 103, 929–938. [Google Scholar] [CrossRef]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and Development of the G-rich Oligonucleotide AS1411 as a Novel Treatment for Cancer. Exp. Mol. Pathol. 2010, 86, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef]
- Chen, Z.; Xu, X.H. Roles of nucleolin: Focus on cancer and anti-cancer therapy. Saudi Med. J. 2016, 37, 1312–1318. [Google Scholar] [CrossRef]
- Corbet, C.; Ragelle, H.; Pourcelle, V.; Vanvarenberg, K.; Marchand-Brynaert, J.; Préat, V.; Feron, O. Delivery of siRNA targeting tumor metabolism using non-covalent PEGylated chitosan nanoparticles: Identification of an optimal combination of ligand structure, linker and grafting method. J. Control. Release 2016, 223, 53–63. [Google Scholar] [CrossRef]
- Ye, S.F.; Tian, M.M.; Wang, T.X.; Ren, L.; Wang, D.; Shen, L.H.; Shang, T. Synergistic effects of cell-penetrating peptide Tat and fusogenic peptide HA2-enhanced cellular internalization and gene transduction of organosilica nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2012, 8, 833–841. [Google Scholar] [CrossRef]
- Li, R.; Peng, F.; Cai, J.; Yang, D.; Zhang, P. Redox dual-stimuli responsive drug delivery systems for improving tumor-targeting ability and reducing adverse side effects. Asian J. Pharm. Sci. 2020, 15, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Hirata, E.; Sahai, E. Tumor microenvironment and differential responses to therapy. Cold Spring Harb. Perspect. Med. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lushchak, V.I. Glutathione Homeostasis and Functions: Potential Targets for Medical Interventions. J. Amino Acids 2012, 2012, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, D.J.; Gibson, M.I. Redox-sensitive materials for drug delivery: Targeting the correct intracellular environment, tuning release rates, and appropriate predictive systems. Antioxidants Redox Signal. 2014, 21, 786–803. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.; Rana, T.M. siRNA function in RNAi: A chemical modification analysis. RNA 2003, 9, 1034–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusconi, C.P.; Scardino, E.; Layzer, J.; Pitoc, G.A.; Ortel, T.L.; Monroe, D.; Sullenger, B.A. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 2002, 419, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Park, K. Synthesis and characterization of superporous hydrogel composites. J. Control. Release 2000, 65, 73–82. [Google Scholar] [CrossRef]
- Shibuya, M. Vascular Endothelial Growth Factor (VEGF) and Its Receptor (VEGFR) Signaling in Angiogenesis: A Crucial Target for Anti- and Pro-Angiogenic Therapies. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef]
- Frokjaer, S.; Otzen, D.E. Protein drug stability: A formulation challenge. Nat. Rev. Drug Discov. 2005, 4, 298–306. [Google Scholar] [CrossRef]
- Coester, C.J.; Langer, K.; van Briesen, H.; Kreuter, J. Gelatin nanoparticles by two step desolvation a new preparation method, surface modifications and cell uptake. J. Microencapsul. 2000, 17, 187–193. [Google Scholar] [CrossRef]
- Nichol, J.W.; Koshy, S.T.; Bae, H.; Hwang, C.M.; Yamanlar, S.; Khademhosseini, A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials 2010, 31, 5536–5544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratanavaraporn, J.; Damrongsakkul, S.; Sanchavanakit, N.; Banaprasert, T.; Kanokpanont, S. Comparison of Gelatin and Collagen Scaffolds for Fibroblast Cell Culture. J. Met. Mater. Miner. 2006, 16, 31–36. [Google Scholar]
- Jongjareonrak, A.; Benjakul, S.; Visessanguan, W.; Nagai, T.; Tanaka, M. Isolation and characterisation of acid and pepsin-solubilised collagens from the skin of Brownstripe red snapper (Lutjanus vitta). Food Chem. 2005, 93, 475–484. [Google Scholar] [CrossRef]
- Krenzlin, H.; Lorenz, V.; Danckwardt, S.; Kempski, O.; Alessandri, B. The importance of thrombin in cerebral injury and disease. Int. J. Mol. Sci. 2016, 17, 84. [Google Scholar] [CrossRef] [Green Version]
- Wolberg, A.S. Thrombin generation and fibrin clot structure. Blood Rev. 2007, 21, 131–142. [Google Scholar] [CrossRef]
- Sweet, R.A.; Nimgaonkar, V.L.; Llyas Kamboh, M.; Lopez, O.L.; Zhang, F.; Dekosky, S.T. Dopamine receptor genetic variation, psychosis, and aggression in Alzheimer disease. Arch. Neurol. 1998, 55, 1335–1340. [Google Scholar] [CrossRef] [Green Version]
- Malapani, C.; Rakitin, B.; Levy, R.; Meck, W.H.; Deweer, B.; Dubois, B.; Gibbon, J. Coupled temporal memories in Parkinson’s disease: A dopamine-related dysfunction. J. Cogn. Neurosci. 1998, 10, 316–331. [Google Scholar] [CrossRef]
- Kasten, M.J. Clindamycin, metronidazole, and chloramphenicol. Mayo Clin. Proc. 1999, 74, 825–833. [Google Scholar] [CrossRef]
- Moriguchi, I.; Hirono, S.; Nagakome, I.; Hirano, H. Comparison of Reliability of log P Values for Drugs Calculated by Several Methods. Chem. Pharm. Bull. (Tokyo) 1994, 42, 976–978. [Google Scholar] [CrossRef] [Green Version]
- Pilehvar, S.; Mehta, J.; Dardenne, F.; Robbens, J.; Blust, R.; De Wael, K. Aptasensing of chloramphenicol in the presence of its analogues: Reaching the maximum residue limit. Anal. Chem. 2012, 84, 6753–6758. [Google Scholar] [CrossRef]
- Vrhovski, B.; Weiss, A.S. Biochemistry of tropoelastin. Eur. J. Biochem. 1998, 258, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Almine, J.F.; Bax, D.V.; Mithieux, S.M.; Smith, L.N.; Rnjak, J.; Waterhouse, A.; Wise, S.G.; Weiss, A.S. Elastin-based materials. Chem. Soc. Rev. 2010, 39, 3371–3379. [Google Scholar] [CrossRef] [PubMed]
- Brown-Augsburger, P.; Broekelmann, T.; Rosenbloom, J.; Mecham, R.P. Functional domains on elastin and microfibril-associated glycoprotein involved in elastic fibre assembly. Biochem. J. 1996, 318, 149–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristensen, J.H.; Karsdal, M.A. Elastin. In Biochemistry of Collagens, Laminins and Elastin; Karsdal, M.A., Ed.; Elsevier: Cambridge, MA, USA, 2016; pp. 197–201. ISBN 9780128098479. [Google Scholar]
- Vindin, H.; Mithieux, S.M.; Weiss, A.S. Elastin architecture. Matrix Biol. 2019, 84, 4–16. [Google Scholar] [CrossRef]
- Urry, D.W.; Cunningham, W.D.; Ohnishi, T. Conformation and Interactions of Elastin. Proton Magnetic Resonance of the Repeating Pentapeptide. Biochemistry 1974, 13, 609–616. [Google Scholar] [CrossRef]
- Mcpherson, D.T.; Morrow, C.; Minehan, D.S.; Wu, J.; Hunter, E.; Urry, D.W. Production and Purification of a Recombinant Elastomeric Polypeptide, G-(VPGVG)19-VPGV, from Escherichia coli. Biotechnol. Prog. 1992, 8, 347–352. [Google Scholar] [CrossRef]
- Won, J.I.; Barron, A.E. A new cloning method for the preparation of long repetitive polypeptides without a sequence requirement. Macromolecules 2002, 35, 8281–8287. [Google Scholar] [CrossRef]
- Urry, D.W. Physical Chemistry of Biological Free Energy Transduction As Demonstrated by Elastic Protein-Based Polymers. J. Phys. Chem. B 1997, 101, 11007–11028. [Google Scholar] [CrossRef]
- Urry, D.W.; Luan, C.H.; Parker, T.M.; Gowda, D.C.; Prasad, K.U.; Reid, M.C.; Safavy, A. Temperature of Polypeptide Inverse Temperature Transition Depends on Mean Residue Hydrophobicity. J. Am. Chem. Soc. 1991, 113, 4346–4348. [Google Scholar] [CrossRef]
- Muiznieks, L.D.; Weiss, A.S.; Keeley, F.W. Structural disorder and dynamics of elastin. Biochem. Cell Biol. 2010, 88, 239–250. [Google Scholar] [CrossRef]
- Tamburro, A.M. A never-ending love story with elastin: A scientific autobiography. Nanomedicine 2009, 4, 469–487. [Google Scholar] [CrossRef] [PubMed]
- MacEwan, S.R.; Chilkoti, A. Applications of elastin-like polypeptides in drug delivery. J. Control. Release 2014, 190, 314–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haugh, M.G.; Vaughan, T.J.; Madl, C.M.; Raftery, R.M.; McNamara, L.M.; O’Brien, F.J.; Heilshorn, S.C. Investigating the interplay between substrate stiffness and ligand chemistry in directing mesenchymal stem cell differentiation within 3D macro-porous substrates. Biomaterials 2018, 171, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Valdivieso, J.; Girotti, A.; Muñoz, R.; Rodriguez-Cabello, J.C.; Arias, F.J. Self-Assembling ELR-Based Nanoparticles as Smart Drug-Delivery Systems Modulating Cellular Growth via Akt. Biomacromolecules 2019, 20, 1996–2007. [Google Scholar] [CrossRef]
- Lee, C.H.; Ingrole, R.S.J.; Gill, H.S. Generation of induced pluripotent stem cells using elastin like polypeptides as a non-viral gene delivery system. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866. [Google Scholar] [CrossRef]
- Salinas-Fernández, S.; Santos, M.; Alonso, M.; Quintanilla, L.; Rodríguez-Cabello, J.C. Genetically engineered elastin-like recombinamers with sequence-based molecular stabilization as advanced bioinks for 3D bioprinting. Appl. Mater. Today 2020, 18, 100500. [Google Scholar] [CrossRef]
- Mahara, A.; Kiick, K.L.; Yamaoka, T. In vivo guided vascular regeneration with a non-porous elastin-like polypeptide hydrogel tubular scaffold. J. Biomed. Mater. Res. Part A 2017, 105, 1746–1755. [Google Scholar] [CrossRef]
- Staubli, S.M.; Cerino, G.; Gonzalez De Torre, I.; Alonso, M.; Oertli, D.; Eckstein, F.; Glatz, K.; Rodríguez Cabello, J.C.; Marsano, A. Control of angiogenesis and host response by modulating the cell adhesion properties of an Elastin-Like Recombinamer-based hydrogel. Biomaterials 2017, 135, 30–41. [Google Scholar] [CrossRef] [Green Version]
- Brody, H. Gene therapy. Nature 2018, 564, S5. [Google Scholar] [CrossRef]
- Hossain, J.A.; Riecken, K.; Miletic, H.; Fehse, B. Cancer Suicide Gene Therapy with TK.007. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2019; Volume 1895, pp. 11–26. [Google Scholar]
- Ginn, S.L.; Amaya, A.K.; Alexander, I.E.; Edelstein, M.; Abedi, M.R. Gene therapy clinical trials worldwide to 2017: An update. J. Gene Med. 2018, 20, e3015. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiriveedhi, V.; Tucker, N.; Herndon, J.; Li, J.; Sturmoski, M.; Ellis, M.; Ma, C.; Naughton, M.; Lockhart, A.C.; Gao, F.; et al. Safety and preliminary evidence of biologic efficacy of a mammaglobin-A DNA vaccine in patients with stable metastatic breast cancer. Clin. Cancer Res. 2014, 20, 5964–5975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vago, R.; Collico, V.; Zuppone, S.; Prosperi, D.; Colombo, M. Nanoparticle-mediated delivery of suicide genes in cancer therapy. Pharmacol. Res. 2016, 111, 619–641. [Google Scholar] [CrossRef] [PubMed]
- Bechara, C.; Sagan, S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013, 587, 1693–1702. [Google Scholar] [CrossRef]
- Blau, S.; Jubeh, T.T.; Haupt, S.M.; Rubinstein, A. Drug Targeting by Surface Cationization. Crit. Rev. Ther. Drug Carr. Syst. 2000, 17, 41. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef]
- Veronesi, U.; Goldhirsch, A.; Veronesi, P.; Gentilini, O.D.; Leonardi, M.C. (Eds.) Breast Cancer; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; ISBN 978-3-319-48846-2. [Google Scholar]
- Piña, M.J.; Alex, S.M.; Arias, F.J.; Santos, M.; Rodríguez-Cabello, J.C.; Ramesan, R.M.; Sharma, C.P. Elastin-like recombinamers with acquired functionalities for gene-delivery applications. J. Biomed. Mater. Res. Part A 2015, 103, 3166–3178. [Google Scholar] [CrossRef] [Green Version]
- Rejman, J.; Oberle, V.; Zuhorn, I.S.; Hoekstra, D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004, 377, 159–169. [Google Scholar] [CrossRef]
- Sahay, G.; Alakhova, D.Y.; Kabanov, A.V. Endocytosis of nanomedicines. J. Control. Release 2010, 145, 182–195. [Google Scholar] [CrossRef] [Green Version]
- Jing, X.; Liang, H.; Hao, C.; Yang, X.; Cui, X. Overexpression of MUC1 predicts poor prognosis in patients with breast cancer. Oncol. Rep. 2019, 41, 801–810. [Google Scholar] [CrossRef]
- Kattula, S.; Byrnes, J.R.; Wolberg, A.S. Fibrinogen and Fibrin in Hemostasis and Thrombosis. Arterioscler. Thromb. Vasc. Biol. 2017, 37, e13–e21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuliani-Alvarez, L.; Midwood, K.S. Fibrinogen-Related Proteins in Tissue Repair: How a Unique Domain with a Common Structure Controls Diverse Aspects of Wound Healing. Adv. Wound Care 2015, 4, 273–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parry, D.A.D.; Squire, J.M. Fibrous Proteins: Structures and Mechanisms; Subcellular Biochemistry; Parry, D.A.D., Squire, J.M., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; ISBN 978-3-319-49672-6. [Google Scholar]
- Weisel, J.W. Fibrinogen and Fibrin. Adv. Protein Chem. 2005, 70, 247–299. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.C.; Barker, T.H. Fibrin-based biomaterials: Modulation of macroscopic properties through rational design at the molecular level. Acta Biomater. 2014, 10, 1502–1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falvo, M.R.; Gorkun, O.V.; Lord, S.T. The molecular origins of the mechanical properties of fibrin. Biophys. Chem. 2010, 152, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Noori, A.; Ashrafi, S.J.; Vaez-Ghaemi, R.; Hatamian-Zaremi, A.; Webster, T.J. A review of fibrin and fibrin composites for bone tissue engineering. Int. J. Nanomed. 2017, 12, 4937–4961. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, T.A.E.; Dare, E.V.; Hincke, M. Fibrin: A versatile scaffold for tissue engineering applications. Tissue Eng. Part B Rev. 2008, 14, 199–215. [Google Scholar] [CrossRef]
- Jockenhoevel, S.; Zund, G.; Hoerstrup, S.P.; Chalabi, K.; Sachweh, J.S.; Demircan, L.; Messmer, B.J.; Turina, M. Fibrin gel—Advantages of a new scaffold in cardiovascular tissue engineering. Eur. J. Cardiothoracic Surg. 2001, 19, 424–430. [Google Scholar] [CrossRef] [Green Version]
- Clegg, J.R.; Wechsler, M.E.; Peppas, N.A. Vision for Functionally Decorated and Molecularly Imprinted Polymers in Regenerative Engineering. Regen. Eng. Transl. Med. 2017, 3, 166–175. [Google Scholar] [CrossRef]
- Lee, K.; Silva, E.A.; Mooney, D.J. Growth factor delivery-based tissue engineering: General approaches and a review of recent developments. J. R. Soc. Interface 2011, 8, 153–170. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, Z.; Lu, W.W.; Zhen, W.; Yang, D.; Peng, S. Novel biomaterial strategies for controlled growth factor delivery for biomedical applications. NPG Asia Mater. 2017, 9, e435. [Google Scholar] [CrossRef]
- Liu, Y.; Hou, W.; Sun, H.; Cui, C.; Zhang, L.; Jiang, Y.; Wu, Y.; Wang, Y.; Li, J.; Sumerlin, B.S.; et al. Thiol-ene click chemistry: A biocompatible way for orthogonal bioconjugation of colloidal nanoparticles. Chem. Sci. 2017, 8, 6182–6187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrara, N.; Gerber, H.P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Brudno, Y.; Ennett-Shepard, A.B.; Chen, R.R.; Aizenberg, M.; Mooney, D.J. Enhancing microvascular formation and vessel maturation through temporal control over multiple pro-angiogenic and pro-maturation factors. Biomaterials 2013, 34, 9201–9209. [Google Scholar] [CrossRef] [Green Version]
- Battig, M.R.; Huang, Y.; Chen, N.; Wang, Y. Aptamer-functionalized superporous hydrogels for sequestration and release of growth factors regulated via molecular recognition. Biomaterials 2014, 35, 8040–8048. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef] [Green Version]
- Street, J.; Bao, M.; DeGuzman, L.; Bunting, S.; Peale, F.V.; Ferrara, N.; Steinmetz, H.; Hoeffel, J.; Cleland, J.L.; Daugherty, A.; et al. Vascular endothelial growth factor stimulates bone repair by promoting angiogenesis and bone turnover. Proc. Natl. Acad. Sci. USA 2002, 99, 9656–9661. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Jahagirdar, B.N.; Reinhardt, R.L.; Schwartz, R.E.; Keene, C.D.; Ortiz-Gonzalez, X.R.; Reyes, M.; Lenvik, T.; Lund, T.; Blackstad, M.; et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 2002, 418, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Pittenger, M.F.; Discher, D.E.; Péault, B.M.; Phinney, D.G.; Hare, J.M.; Caplan, A.I. Mesenchymal stem cell perspective: Cell biology to clinical progress. npj Regen. Med. 2019, 4, 22. [Google Scholar] [CrossRef] [Green Version]
- Hmadcha, A.; Martin-Montalvo, A.; Gauthier, B.R.; Soria, B.; Capilla-Gonzalez, V. Therapeutic Potential of Mesenchymal Stem Cells for Cancer Therapy. Front. Bioeng. Biotechnol. 2020, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, X.; Wang, W.E.; Zeng, C. How to Improve the Survival of Transplanted Mesenchymal Stem Cell in Ischemic Heart? Stem Cells Int. 2016, 2, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frith, J.E.; Thomson, B.; Genever, P.G. Dynamic three-dimensional culture methods enhance mesenchymal stem cell properties and increase therapeutic potential. Tissue Eng. Part C Methods 2010, 16, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Im, J.H.; Fu, W.; Wang, H.; Bhatia, S.K.; Hammer, D.A.; Kowalska, M.A.; Muschel, R.J. Coagulation facilitates tumor cell spreading in the pulmonary vasculature during early metastatic colony formation. Cancer Res. 2004, 64, 8613–8619. [Google Scholar] [CrossRef] [Green Version]
- Idell, S.; Pueblitz, S.; Emri, S.; Gungen, Y.; Gray, L.; Kumar, A.; Holiday, D.; Koenig, K.B.; Johnson, A.R. Regulation of fibrin deposition by malignant mesothelioma. Am. J. Pathol. 1995, 147, 1318–1329. [Google Scholar]
- Dvorak, H.F.; Senger, D.R.; Dvorak, A.M. Fibrin as a component of the tumor stroma: Origins and biological significance. Cancer Metastasis Rev. 1983, 2, 41–73. [Google Scholar] [CrossRef]
- Fujita, H.; Nakajima, K.; Kasahara, Y.; Ozaki, H.; Kuwahara, M. Polymerase-mediated high-density incorporation of amphiphilic functionalities into DNA: Enhancement of nuclease resistance and stability in human serum. Bioorg. Med. Chem. Lett. 2015, 25, 333–336. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, H.; Kasahara, Y.; Fujita, H.; Kuwahara, M.; Morihiro, K.; Tsunoda, S.I.; Obika, S. Consecutive incorporation of functionalized nucleotides with amphiphilic side chains by novel KOD polymerase mutant. Bioorg. Med. Chem. Lett. 2016, 26, 530–533. [Google Scholar] [CrossRef]
- Nyga, A.; Cheema, U.; Loizidou, M. 3D tumour models: Novel in vitro approaches to cancer studies. J. Cell Commun. Signal. 2011, 5, 239–248. [Google Scholar] [CrossRef] [Green Version]
- Westhouse, R.A. Safety assessment considerations and strategies for targeted small molecule cancer therapeutics in drug discovery. Toxicol. Pathol. 2010, 38, 165–168. [Google Scholar] [CrossRef]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083. [Google Scholar] [CrossRef]
- Wall, M.E.; Wani, M.C.; Cook, C.E.; Palmer, K.H.; McPhail, A.T.; Sim, G.A. Plant Antitumor Agents. I. The Isolation and Structure of Camptothecin, a Novel Alkaloidal Leukemia and Tumor Inhibitor from Camptotheca acuminata. J. Am. Chem. Soc. 1966, 88, 3888–3890. [Google Scholar] [CrossRef]
- Li, F.; Jiang, T.; Li, Q.; Ling, X. Camptothecin analogues and their molecular targets. Am. J. Cancer Res. 2017, 7, 2350–2394. [Google Scholar] [PubMed]
- Huang, W.; Ling, S.; Li, C.; Omenetto, F.G.; Kaplan, D.L. Silkworm silk-based materials and devices generated using bio-nanotechnology. Chem. Soc. Rev. 2018, 47, 6486–6504. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, P.; Kundu, B.; Naskar, D.; Kim, H.-W.; Maiti, T.K.; Bhattacharya, D.; Kundu, S.C. Silk scaffolds in bone tissue engineering: An overview. Acta Biomater. 2017, 63, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chawla, S.; Midha, S.; Sharma, A.; Ghosh, S. Silk-Based Bioinks for 3D Bioprinting. Adv. Healthc. Mater. 2018, 7, 1701204. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, T.D.; Young, J.H.; Weisman, S.; Hayashi, C.Y.; Merritt, D.J. Insect Silk: One Name, Many Materials. Annu. Rev. Entomol. 2010, 55, 171–188. [Google Scholar] [CrossRef] [PubMed]
- Altman, G.H.; Diaz, F.; Jakuba, C.; Calabro, T.; Horan, R.L.; Chen, J.; Lu, H.; Richmond, J.; Kaplan, D.L. Silk-based biomaterials. Biomaterials 2003, 24, 401–416. [Google Scholar] [CrossRef] [Green Version]
- Vepari, C.; Kaplan, D.L. Silk as a biomaterial. Prog. Polym. Sci. 2007, 32, 991–1007. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Khademhosseini, A. Advances in engineering hydrogels. Science 2017, 356, eaaf3627. [Google Scholar] [CrossRef]
- Pica, A.; Krauss, I.R.; Parente, V.; Tateishi-Karimata, H.; Nagatoishi, S.; Tsumoto, K.; Sugimoto, N.; Sica, F. Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers. Nucleic Acids Res. 2017, 45, 461–469. [Google Scholar] [CrossRef] [Green Version]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiang, Y.-C.; Huang, C.-C.; Wang, T.-H.; Chien, C.-W.; Chang, H.-T. Aptamer-Conjugated Nanoparticles Efficiently Control the Activity of Thrombin. Adv. Funct. Mater. 2010, 20, 3175–3182. [Google Scholar] [CrossRef]
- Benvidi, A.; Banaei, M.; Tezerjani, M.D.; Molahosseini, H.; Jahanbani, S. Impedimetric PSA aptasensor based on the use of a glassy carbon electrode modified with titanium oxide nanoparticles and silk fibroin nanofibers. Microchim. Acta 2018, 185, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sivaraman, A.; Bhat, K.R.S. Screening and Detection of Prostate Cancer—Review of Literature and Current Perspective. Indian J. Surg. Oncol. 2017, 8, 160–168. [Google Scholar] [CrossRef]
- Marrazza, G. Aptamer Sensors. Biosensors 2017, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Lapčík, L.; Lapčík, L.; De Smedt, S.; Demeester, J.; Chabreček, P. Hyaluronan: Preparation, structure, properties, and applications. Chem. Rev. 1998, 98. [Google Scholar] [CrossRef]
- Toole, B.P. Hyaluronan: From extracellular glue to pericellular cue. Nat. Rev. Cancer 2004, 4, 528–539. [Google Scholar] [CrossRef]
- Lozoya, O.A.; Wauthier, E.; Turner, R.A.; Barbier, C.; Prestwich, G.D.; Guilak, F.; Superfine, R.; Lubkin, S.R.; Reid, L.M. Regulation of hepatic stem/progenitor phenotype by microenvironment stiffness in hydrogel models of the human liver stem cell niche. Biomaterials 2011, 32, 7389–7402. [Google Scholar] [CrossRef] [Green Version]
- Borzacchiello, A.; Russo, L.; Malle, B.M.; Schwach-Abdellaoui, K.; Ambrosio, L. Hyaluronic acid based hydrogels for regenerative medicine applications. Biomed Res. Int. 2015, 2015. [Google Scholar] [CrossRef]
- Neuman, M.G.; Nanau, R.M.; Oruña-Sanchez, L.; Coto, G. Hyaluronic acid and wound healing. J. Pharm. Pharm. Sci. 2015, 18, 53–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volpi, N.; Schiller, J.; Stern, R.; Soltes, L. Role, Metabolism, Chemical Modifications and Applications of Hyaluronan. Curr. Med. Chem. 2009, 16, 1718–1745. [Google Scholar] [CrossRef] [PubMed]
- Allison, D.D.; Grande-Allen, K.J. Review. Hyaluronan: A Powerful Tissue Engineering Tool. Tissue Eng. 2006, 12, 2131–2140. [Google Scholar] [CrossRef] [PubMed]
- Eng, D.; Caplan, M.; Preul, M.; Panitch, A. Hyaluronan scaffolds: A balance between backbone functionalization and bioactivity. Acta Biomater. 2010, 6, 2407–2414. [Google Scholar] [CrossRef]
- Caicco, M.J.; Zahir, T.; Mothe, A.J.; Ballios, B.G.; Kihm, A.J.; Tator, C.H.; Shoichet, M.S. Characterization of hyaluronan-methylcellulose hydrogels for cell delivery to the injured spinal cord. J. Biomed. Mater. Res. Part A 2013, 101A, 1472–1477. [Google Scholar] [CrossRef]
- Knopf-Marques, H.; Pravda, M.; Wolfova, L.; Velebny, V.; Schaaf, P.; Vrana, N.E.; Lavalle, P. Hyaluronic Acid and Its Derivatives in Coating and Delivery Systems: Applications in Tissue Engineering, Regenerative Medicine and Immunomodulation. Adv. Healthc. Mater. 2016, 5, 2841–2855. [Google Scholar] [CrossRef]
- Raia, N.R.; Partlow, B.P.; McGill, M.; Kimmerling, E.P.; Ghezzi, C.E.; Kaplan, D.L. Enzymatically crosslinked silk-hyaluronic acid hydrogels. Biomaterials 2017, 131, 58–67. [Google Scholar] [CrossRef]
- Pereira, H.; Sousa, D.A.; Cunha, A.; Andrade, R.; Espregueira-Mendes, J.; Oliveira, J.M.; Reis, R.L. Hyaluronic acid. In Advances in Experimental Medicine and Biology; Springer LLC.: New York, NY, USA, 2018; Volume 1059, pp. 137–153. [Google Scholar]
- Huayamares, S.G.; Song, J.Y.; Huang, A.; Crowl, S.R.; Groer, C.E.; Forrest, M.L.; Berkland, C.J. Constructing a Biomaterial to Simulate Extracellular Drug Transport in Solid Tumors. Macromol. Biosci. 2020, 2000251. [Google Scholar] [CrossRef]
- Shu, X.Z.; Liu, Y.; Palumbo, F.S.; Luo, Y.; Prestwich, G.D. In situ crosslinkable hyaluronan hydrogels for tissue engineering. Biomaterials 2004, 25, 1339–1348. [Google Scholar] [CrossRef]
- Prestwich, G.D. Hyaluronic acid-based clinical biomaterials derived for cell and molecule delivery in regenerative medicine. J. Control. Release 2011, 155, 193–199. [Google Scholar] [CrossRef] [Green Version]
- Hemshekhar, M.; Thushara, R.M.; Chandranayaka, S.; Sherman, L.S.; Kemparaju, K.; Girish, K.S. Emerging roles of hyaluronic acid bioscaffolds in tissue engineering and regenerative medicine. Int. J. Biol. Macromol. 2016, 86, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fan, D. Novel hyaluronic acid-tyrosine/collagen-based injectable hydrogels as soft filler for tissue engineering. Int. J. Biol. Macromol. 2019, 141, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Shin, M.; Han, S.; Kwon, W.; Hahn, S.K. Hyaluronic Acid Derivatives for Translational Medicines. Biomacromolecules 2019, 20, 2889–2903. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Bahadur, P. Modified hyaluronic acid based materials for biomedical applications. Int. J. Biol. Macromol. 2019, 121, 556–571. [Google Scholar] [CrossRef]
- Ahmadian, E.; Dizaj, S.M.; Eftekhari, A.; Dalir, E.; Vahedi, P.; Hasanzadeh, A.; Samiei, M. The Potential Applications of Hyaluronic Acid Hydrogels in Biomedicine. Drug Res. (Stuttg.) 2020, 70, 6–11. [Google Scholar] [CrossRef]
- Zhu, X.; Zhou, H.; Liu, Y.; Wen, Y.; Wei, C.; Yu, Q.; Liu, J. Transferrin/aptamer conjugated mesoporous ruthenium nanosystem for redox-controlled and targeted chemo-photodynamic therapy of glioma. Acta Biomater. 2018, 82, 143–157. [Google Scholar] [CrossRef]
- Gongol, B.; Marin, T.; Zhang, J.; Wang, S.C.; Sun, W.; He, M.; Chen, S.; Chen, L.; Li, J.; Liu, J.H.; et al. Shear stress regulation of miR-93 and miR-484 maturation through nucleolin. Proc. Natl. Acad. Sci. USA 2019, 116, 12974–12979. [Google Scholar] [CrossRef] [Green Version]
- Farokhzad, O.C.; Karp, J.M.; Langer, R. Nanoparticle–aptamer bioconjugates for cancer targeting. Expert Opin. Drug Deliv. 2006, 3, 311–324. [Google Scholar] [CrossRef]
- Brittis, P.A.; Flanagan, J.G. Nogo Domains and a Nogo Receptor:Implications for Axon Regeneration. Neuron 2001, 30, 11–14. [Google Scholar] [CrossRef] [Green Version]
- Fournier, A.E.; GrandPre, T.; Strittmatter, S.M. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature 2001, 409, 341–346. [Google Scholar] [CrossRef]
- Wang, Y.; Khaing, Z.Z.; Li, N.; Hall, B.; Schmidt, C.E.; Ellington, A.D. Aptamer Antagonists of Myelin-Derived Inhibitors Promote Axon Growth. PLoS ONE 2010, 5, e9726. [Google Scholar] [CrossRef] [PubMed]
- Highley, C.B.; Prestwich, G.D.; Burdick, J.A. Recent advances in hyaluronic acid hydrogels for biomedical applications. Curr. Opin. Biotechnol. 2016, 40, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Di Buduo, C.A.; Abbonante, V.; Tozzi, L.; Kaplan, D.L.; Balduini, A. Three-Dimensional Tissue Models for Studying Ex Vivo Megakaryocytopoiesis and Platelet Production. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2018; Volume 1812, pp. 177–193. [Google Scholar]
- Raia, N.R.; Jia, D.; Ghezzi, C.E.; Muthukumar, M.; Kaplan, D.L. Characterization of silk-hyaluronic acid composite hydrogels towards vitreous humor substitutes. Biomaterials 2020, 233. [Google Scholar] [CrossRef] [PubMed]
- Yodmuang, S.; McNamara, S.L.; Nover, A.B.; Mandal, B.B.; Agarwal, M.; Kelly, T.A.N.; Chao, P.H.G.; Hung, C.; Kaplan, D.L.; Vunjak-Novakovic, G. Silk microfiber-reinforced silk hydrogel composites for functional cartilage tissue repair. Acta Biomater. 2015, 11, 27–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toh, W.S.; Lim, T.C.; Kurisawa, M.; Spector, M. Modulation of mesenchymal stem cell chondrogenesis in a tunable hyaluronic acid hydrogel microenvironment. Biomaterials 2012, 33, 3835–3845. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.L.; Khetan, S.; Baker, B.M.; Chen, C.S.; Burdick, J.A. Fibrous hyaluronic acid hydrogels that direct MSC chondrogenesis through mechanical and adhesive cues. Biomaterials 2013, 34, 5571–5580. [Google Scholar] [CrossRef] [Green Version]
- Rapuano, B.E.; Hackshaw, K.M.; Schniepp, H.C.; MacDonald, D.E. Effects of coating a titanium alloy with fibronectin on the expression of osteoblast gene markers in the MC3T3 osteoprogenitor cell line. Int. J. Oral Maxillofac. Implants 2012, 27, 1081–1090. [Google Scholar] [PubMed]
- Pendegrass, C.J.; El-Husseiny, M.; Blunn, G.W. The development of fibronectin-functionalised hydroxyapatite coatings to improve dermal fibroblast attachment in vitro. J. Bone Jt. Surg. Ser. B 2012, 94 B, 564–569. [Google Scholar] [CrossRef]
- Chatakun, P.; Núñez-Toldrà, R.; Díaz López, E.J.; Gil-Recio, C.; Martínez-Sarrà, E.; Hernández-Alfaro, F.; Ferrés-Padró, E.; Giner-Tarrida, L.; Atari, M. The effect of five proteins on stem cells used for osteoblast differentiation and proliferation: A current review of the literature. Cell. Mol. Life Sci. 2014, 71, 113–142. [Google Scholar] [CrossRef]
- Vinsova, J.; Vavrikova, E. Chitosan Derivatives with Antimicrobial, Antitumour and Antioxidant Activities—A Review. Curr. Pharm. Des. 2011, 17, 3596–3607. [Google Scholar] [CrossRef]
- Muzzarelli, R.A.A.; Greco, F.; Busilacchi, A.; Sollazzo, V.; Gigante, A. Chitosan, hyaluronan and chondroitin sulfate in tissue engineering for cartilage regeneration: A review. Carbohydr. Polym. 2012, 89, 723–739. [Google Scholar] [CrossRef] [PubMed]
- Mok, H.; Ji, W.P.; Tae, G.P. Antisense oligodeoxynucleotide-conjugated hyaluronic acid/protamine nanocomplexes for intracellular gene inhibition. Bioconjug. Chem. 2007, 18, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Kufe, D.W. MUC1-C oncoprotein as a target in breast cancer: Activation of signaling pathways and therapeutic approaches. Oncogene 2013, 32, 1073–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebrahimnejad, P.; Dinarvand, R.; Sajadi, A.; Jaafari, M.R.; Nomani, A.R.; Azizi, E.; Rad-Malekshahi, M.; Atyabi, F. Preparation and in vitro evaluation of actively targetable nanoparticles for SN-38 delivery against HT-29 cell lines. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Kawato, Y.; Aonuma, M.; Hirota, Y.; Kuga, H.; Sato, K. Intracellular Roles of SN-38, a Metabolite of the Camptothecin Derivative CPT-11, in the Antitumor Effect of CPT-11. Cancer Res. 1991, 51, 4187–4191. [Google Scholar] [PubMed]








| Biomaterial | Scaffold | Type of Aptamer | Target | Application | Reference |
|---|---|---|---|---|---|
| Atelocollagen | Complex | RNA | PSMA | Prostate cancer derived bone metastasis | [20] |
| Gelatin-Silica + PEG | Nanoparticles | DNA | Nucleolin | Gene delivery | [21] |
| Gelatin-Silica | Nanogels | DNA | Nucleolin | siRNA delivery | [22] |
| Gelatin | Composite | DNA | PDGF-BB | Molecule release | [23] |
| Gelatin-PEG | Hydrogel | RNA | VEGF | Cell and Growth factor sequestration | [24] |
| Gelatin | Nanoparticle assembly hydrogel | RNA | VEGF | Growth Factor sequestration and release | [25] |
| Collagen | Coating | DNA | Thrombin | Biosensing | [26,27] |
| Collagen-Graphene oxide | Composite | DNA | Dopamine | Biosensing | [28] |
| Gelatin | Coating | DNA | Chloramphenicol | Biosensing | [29] |
| Elastin | Polyplexes | DNA | MUC-1 | Drug delivery | [30] |
| Elastin and PEG | Polyplexes | DNA | MUC-1 | Drug delivery | [31] |
| Elastin and poly(aspartic acid) | Nanoparticles | DNA | MUC-1 | Drug delivery | [32] |
| Elastin | Nanoparticles | DNA | MUC-1 | Drug delivery | [33] |
| Elastin | Vesicles | RNA | DFHBI molecule | Visualization of cell-free gene expression | [34] |
| Elastin | Giant peptide vesicles | RNA | DFHBI molecule | Visualization of cell-free gene expression | [35] |
| Silk | Nanohydrogel | DNA | Thrombine exosite I and II | Selective and reversible inhibition of thrombin | [36] |
| Fibrin | Hydrogel | DNA | VEGF | Wound healing | [37] |
| Fibrin | Hydrogel | DNA | VEGF and PDGF-BB | Angiogenesis | [38] |
| Fibrin | Hydrogel | DNA | VEGF | Bone healing | [39] |
| Fibrin | Hydrogel | VEGF and PDGF-BB | MSC survival enhancement | [40] | |
| Fibrin | Hydrogel | DNA | Thrombin | Molecule entrapment | [41] |
| Fibrin | Hydrogel | DNA | Thrombin + Camptothecin | Chemotherapy | [42] |
| HA | Microemulsion | DNA | Nucleolin | Glioma | [43] |
| Hydrogel | RNA | NgR | Spinal cord injury | [44] | |
| SF + HA | Hydrogel | DNA | MSCs | Cartilage repair | [45] |
| PEGDA + tHA | Hydrogel | DNA | Fibronectin | Tissue regeneration | [46] |
| HA + Chitosan | Nanoparticle | DNA | MUC1 | Colorectal adenocarcinoma | [47] |
| Nanoparticle | DNA | MUC1 | Chemotherapy | [48] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Girotti, A.; Escalera-Anzola, S.; Alonso-Sampedro, I.; González-Valdivieso, J.; Arias, F.J. Aptamer-Functionalized Natural Protein-Based Polymers as Innovative Biomaterials. Pharmaceutics 2020, 12, 1115. https://doi.org/10.3390/pharmaceutics12111115
Girotti A, Escalera-Anzola S, Alonso-Sampedro I, González-Valdivieso J, Arias FJ. Aptamer-Functionalized Natural Protein-Based Polymers as Innovative Biomaterials. Pharmaceutics. 2020; 12(11):1115. https://doi.org/10.3390/pharmaceutics12111115
Chicago/Turabian StyleGirotti, Alessandra, Sara Escalera-Anzola, Irene Alonso-Sampedro, Juan González-Valdivieso, and Francisco Javier Arias. 2020. "Aptamer-Functionalized Natural Protein-Based Polymers as Innovative Biomaterials" Pharmaceutics 12, no. 11: 1115. https://doi.org/10.3390/pharmaceutics12111115