Perturbative-Iterative Computation of Inertial Manifolds of Systems of Delay-Differential Equations with Small Delays
Abstract
:1. Introduction
2. Iterative Computation of Attracting Invariant Manifolds of ODEs
3. Computation of Inertial Manifolds for DDEs
3.1. The ”Fast RNA” Case
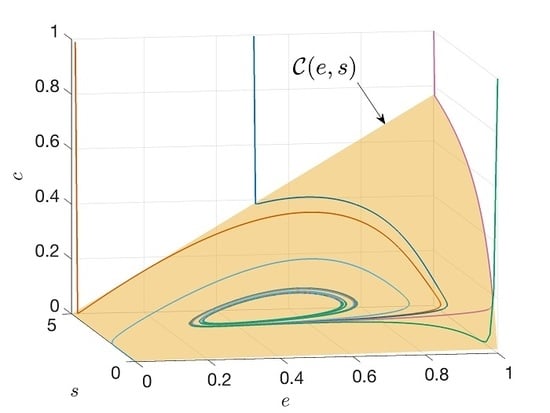
- Because the manifolds computed for different values of m are essentially identical, it appears that the higher-order corrections to the vector field are unnecessary to capture the dynamics, at least in the parameter regime of Figure 4.
- The calculation for (even if carried out numerically) is much better behaved than for larger values of m. In the numerical calculations with larger m, localized imperfections appear in the manifolds. A simple routine was developed for removing these imperfections, essentially by interpolating through the region where they appear, but we can see from Figure 4b that there are small residual artifacts in the surfaces. (, on the other hand, is perfectly smooth.)
- If we integrate the model reduced to the inertial manifold, the reduced model best reproduces the trajectories of the three-variable DDE model (Figure 5). After an unphysical excursion to negative concentrations, the model reproduces the trajectory of the three-variable model reasonably well. On the other hand, the model has a limit cycle, which does not appear in the three-variable model until is above 0.54, and the model is excessively damped. The negative concentrations in the reduced model may be due to a poor representation of the inertial manifold near the axis, and would be an interesting subject for future investigations. The more serious misbehavior of the and models is likely due to a compounding of numerical issues, mostly the difficulties in solving the functional equations accurately in certain parts of the surface, numerical error during the inversion of the relation , and of course the interaction of these two factors.
3.2. Elimination of the Fastest Relaxation Mode
3.3. Computing a Two-Dimensional Inertial Manifold
4. Discussion
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Roussel, M.R.; Fraser, S.J. On the Geometry of Transient Relaxation. J. Chem. Phys. 1991, 94, 7106–7113. [Google Scholar] [CrossRef]
- Lam, S.H. Using CSP to Understand Complex Chemical Kinetics. Combust. Sci. Technol. 1993, 89, 375–404. [Google Scholar] [CrossRef]
- Roussel, M.R.; Fraser, S.J. Invariant Manifold Methods for Metabolic Model Reduction. Chaos 2001, 11, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Lubich, C.; Nipp, K.; Stoffer, D. Runge-Kutta Solutions of Stiff Differential Equations Near Stationary Points. SIAM J. Numer. Anal. 1995, 32, 1296–1307. [Google Scholar] [CrossRef] [Green Version]
- Fraser, S.J. The Steady State and Equilibrium Approximations: A Geometrical Picture. J. Chem. Phys. 1988, 88, 4732–4738. [Google Scholar] [CrossRef]
- Roussel, M.R. Nonlinear Dynamics: A Hands-On Introductory Survey; IOP Concise Physics, Morgan & Claypool: San Rafael, CA, USA, 2019. [Google Scholar] [CrossRef]
- Roberts, A.J. The Utility of an Invariant Manifold Description of the Evolution of a Dynamical System. SIAM J. Math. Anal. 1989, 20, 1447–1458. [Google Scholar] [CrossRef]
- Hale, J.K.; Koçak, H. Dynamics and Bifurcations; Texts in Applied Mathematics; Springer: New York, NY, USA, 1991; Volume 3. [Google Scholar]
- Carr, J. Applications of Centre Manifold Theory; Springer: New York, NY, USA, 1982. [Google Scholar]
- Bogoliubov, N.N.; Mitropolsky, Y.A. The Method of Integral Manifolds in Nonlinear Mechanics. Contrib. Differ. Equ. 1963, 2, 123–196. [Google Scholar]
- Fenichel, N. Persistence and Smoothness of Invariant Manifolds for Flows. Indiana U. Math. J. 1971, 21, 193–226. [Google Scholar] [CrossRef]
- Fenichel, N. Geometric Singular Perturbation Theory for Ordinary Differential Equations. J. Differ. Equ. 1979, 31, 53–98. [Google Scholar] [CrossRef] [Green Version]
- Bliss, R.D. Analysis of the Dynamic Behavior of the Tryptophan Operon of Escherichia coli: The Functional Significance of Feedback Inhibition. Ph.D. Thesis, University of California Riverside, Riverside, CA, USA, 1979. [Google Scholar]
- Mahaffy, J.M. Cellular Control Models with Linked Positive and Negative Feedback and Delays. I. The Models. J. Theor. Biol. 1984, 106, 89–102. [Google Scholar] [CrossRef]
- Lewis, J. Autoinhibition with Transcriptional Delay: A Simple Mechanism for the Zebrafish Somitogenesis Oscillator. Curr. Biol. 2003, 13, 1398–1408. [Google Scholar] [CrossRef] [Green Version]
- Monk, N.A.M. Oscillatory Expression of Hes1, p53, and NF-κB Driven by Transcriptional Time Delays. Curr. Biol. 2003, 13, 1409–1413. [Google Scholar] [CrossRef] [Green Version]
- Ahsen, M.E.; Özbay, H.; Niculescu, S.I. Analysis of Deterministic Cyclic Gene Regulatory Network Models with Delays; Birkhäuser: Cham, Switzerland, 2010. [Google Scholar] [CrossRef]
- Mackey, M.C.; Santillán, M.; Tyran-Kamińska, M.; Zeron, E.S. Simple Mathematical Models of Gene Regulatory Dynamics; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Hale, J.K.; Lunel, S.M.V. Introduction to Functional Differential Equations; Springer: New York, NY, USA, 1993. [Google Scholar]
- Hale, J.K. Dynamics and Delays. In Delay Differential Equations and Dynamical Systems; Busenberg, S., Martelli, M., Eds.; Springer: Berlin, Germany, 1990; pp. 16–30. [Google Scholar]
- Luskin, M.; Sell, G.R. Approximation Theories for Inertial Manifolds. Math. Model. Numer. Anal. 1989, 23, 445–461. [Google Scholar] [CrossRef]
- Fernández, A. The Steady-state Approximation as a Centre Manifold Elimination in Chemical Kinetics. J. Chem. Soc. Faraday Trans. 1986, 82, 849–855. [Google Scholar] [CrossRef]
- Fabes, E.; Luskin, M.; Sell, G.R. Construction of Inertial Manifolds by Elliptic Regularization. J. Differ. Equ. 1991, 89, 355–387. [Google Scholar] [CrossRef] [Green Version]
- Bélair, J.; Campbell, S.A. Similarity and Bifurcations of Equilibria in a Multiple-Delayed Differential Equation. SIAM J. Appl. Math. 1994, 54, 1402–1424. [Google Scholar] [CrossRef]
- Wischert, W.; Wunderlin, A.; Pelster, A.; Olivier, M.; Groslambert, J. Delay-Induced Instabilities in Nonlinear Feedback Systems. Phys. Rev. E 1994, 49, 203–219. [Google Scholar] [CrossRef]
- Qesmi, R.; Ait Babram, M.; Hbid, M.L. A Maple Program for Computing a Terms of a Center Manifold, and Element of Bifurcations for a Class of Retarded Functional Differential Equations with Hopf Singularity. Appl. Math. Comput. 2006, 175, 932–968. [Google Scholar] [CrossRef]
- Campbell, S.A. Calculating Centre Manifolds for Delay Differential Equations Using MapleTM. In Delay Differential Equations: Recent Advances and New Directions; Balachandran, B., Kalmár-Nagy, T., Gilsinn, D.E., Eds.; Springer: New York, NY, USA, 2009; pp. 221–244. [Google Scholar]
- Farkas, G. Unstable Manifolds for RFDEs under Discretization: The Euler Method. Comput. Math. Appl. 2001, 42, 1069–1081. [Google Scholar] [CrossRef] [Green Version]
- Gear, C.W.; Kaper, T.J.; Kevrekidis, I.G.; Zagaris, A. Projecting to a Slow Manifold: Singularly Perturbed Systems and Legacy Codes. SIAM J. Appl. Dyn. Syst. 2005, 4, 711–732. [Google Scholar] [CrossRef] [Green Version]
- Krauskopf, B.; Green, K. Computing Unstable Manifolds of Periodic Orbits in Delay Differential Equations. J. Comput. Phys. 2003, 186, 230–249. [Google Scholar] [CrossRef]
- Green, K.; Krauskopf, B.; Engelborghs, K. One-Dimensional Unstable Eigenfunction and Manifold Computations in Delay Differential Equations. J. Comput. Phys. 2004, 197, 86–98. [Google Scholar] [CrossRef] [Green Version]
- Sahai, T.; Vladimirsky, A. Numerical Methods for Approximating Invariant Manifolds of Delayed Systems. SIAM J. Appl. Dyn. Syst. 2009, 8, 1116–1135. [Google Scholar] [CrossRef] [Green Version]
- Chicone, C. Inertial and Slow Manifolds for Delay Equations with Small Delays. J. Differ. Equ. 2003, 190, 364–406. [Google Scholar] [CrossRef] [Green Version]
- Chekroun, M.D.; Ghil, M.; Liu, H.; Wang, S. Low-Dimensional Galerkin Approximations of Nonlinear Delay Differential Equations. Discret. Contin. Dyn. Syst. 2016, 36, 4133–4177. [Google Scholar] [CrossRef]
- Cunningham, W.J. A Nonlinear Differential-Difference Equation of Growth. Proc. Natl. Acad. Sci. USA 1954, 40, 708–713. [Google Scholar] [CrossRef] [Green Version]
- El’sgol’ts, L.L. Differential Equations; Hindustan: Delhi, India, 1961. [Google Scholar]
- Burke, W.L. Runaway Solutions: Remarks on the Asymptotic Theory of Radiation Damping. Phys. Rev. A 1970, 2, 1501–1505. [Google Scholar] [CrossRef]
- Chicone, C.; Kopeikin, S.M.; Mashhoon, B.; Retzloff, D.G. Delay Equations and Radiation Damping. Phys. Lett. A 2001, 285, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Roussel, M.R. Approximating State-Space Manifolds which Attract Solutions of Systems of Delay-Differential Equations. J. Chem. Phys. 1998, 109, 8154–8160. [Google Scholar] [CrossRef]
- Chicone, C. Inertial Flows, Slow Flows, and Combinatorial Identities for Delay Equations. J. Dyn. Differ. Equ. 2004, 16, 805–831. [Google Scholar] [CrossRef] [Green Version]
- Tomlin, A.S.; Turányi, T.; Pilling, M.J. Mathematical Tools for the Construction, Investigation and Reduction of Combustion Mechanisms. Compr. Chem. Kinet. 1997, 35, 293–437. [Google Scholar]
- Okino, M.S.; Mavrovouniotis, M.L. Simplification of Mathematical Models of Chemical Reaction Systems. Chem. Rev. 1998, 98, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Gorban, A.N.; Karlin, I.V.; Zinovyev, A.Y. Constructive Methods of Invariant Manifolds for Kinetic Problems. Phys. Rep. 2004, 396, 197–403. [Google Scholar] [CrossRef]
- Gorban, A.N.; Karlin, I.V. Invariant Manifolds for Physical and Chemical Kinetics; Lecture Notes in Physics; Springer: Heidelberg, Germany, 2005; Volume 660. [Google Scholar] [CrossRef] [Green Version]
- Chiavazzo, E.; Gorban, A.N.; Karlin, I.V. Comparison of Invariant Manifolds for Model Reduction in Chemical Kinetics. Commun. Comput. Phys. 2007, 2, 964–992. [Google Scholar]
- Maas, U.; Tomlin, A.S. Time-Scale Splitting-Based Mechanism Reduction. In Cleaner Combustion; Battin-Leclerc, F., Simmie, J.M., Blurock, E., Eds.; Springer: London, UK, 2013; Chapter 18; pp. 467–484. [Google Scholar]
- Gorban, A.N. Model Reduction in Chemical Dynamics: Slow Invariant Manifolds, Singular Perturbations, Thermodynamic Estimates, and Analysis of Reaction Graph. Curr. Opin. Chem. Eng. 2018, 21, 48–59. [Google Scholar] [CrossRef] [Green Version]
- Roussel, M.R.; Fraser, S.J. Geometry of the Steady-State Approximation: Perturbation and Accelerated Convergence Methods. J. Chem. Phys. 1990, 93, 1072–1081. [Google Scholar] [CrossRef]
- Roussel, M.R. Forced-Convergence Iterative Schemes for the Approximation of Invariant Manifolds. J. Math. Chem. 1997, 21, 385–393. [Google Scholar] [CrossRef]
- Davis, M.J.; Skodje, R.T. Geometric Investigation of Low-Dimensional Manifolds in Systems Approaching Equilibrium. J. Chem. Phys. 1999, 111, 859–874. [Google Scholar] [CrossRef]
- Engelborghs, K.; Luzyanina, T.; Roose, D. Numerical Bifurcation Analysis of Delay Differential Equations Using DDE-BIFTOOL. ACM Trans. Math. Softw. 2002, 28, 1–21. [Google Scholar] [CrossRef]
- Roussel, M.R. Slowly Reverting Enzyme Inactivation: A Mechanism for Generating Long-lived Damped Oscillations. J. Theor. Biol. 1998, 195, 233–244. [Google Scholar] [CrossRef]
- Roussel, M.R.; Fraser, S.J. Accurate Steady-State Approximations: Implications for Kinetics Experiments and Mechanism. J. Phys. Chem. 1991, 95, 8762–8770. [Google Scholar] [CrossRef]
- Monod, J.; Torriani, A.M.; Gribetz, J. Sur une lactase extraite d’une souche d’Escherichia coli mutabile. C. R. Acad. Sci. 1948, 227, 315–316. [Google Scholar]
- Monod, J.; Pappenheimer, A.M., Jr.; Cohen-Bazire, G. La cinétique de la biosynthèse de la β-galactosidase chez E. coli considérée comme fonction de la croissance. Biochim. Biophys. Acta 1952, 9, 648–660. [Google Scholar] [CrossRef]
- Gardner, P.R.; Gardner, A.M.; Martin, L.A.; Salzman, A.L. Nitric Oxide Dioxygenase: An Enzymic Function for Flavohemoglobin. Proc. Natl. Acad. Sci. USA 1998, 95, 10378–10383. [Google Scholar] [CrossRef] [Green Version]
- Poole, R.K.; Anjum, M.F.; Membrillo-Hernández, J.; Kim, S.O.; Hughes, M.N.; Stewart, V. Nitric Oxide, Nitrite, and Fnr Regulation of hmp (Flavohemoglobin) Gene Expression in Escherichia coli K-12. J. Bacteriol. 1996, 178, 5487–5492. [Google Scholar] [CrossRef] [Green Version]
- Ferrell, J.E., Jr. Tripping the Switch Fantastic: How a Protein Kinase Can Convert Graded Inputs into Switch-Like Outputs. Trends Biochem. Sci. 1996, 21, 460–466. [Google Scholar] [CrossRef]
- Ferrell, J.E., Jr. How Regulated Protein Translocation Can Produce Switch-Like Responses. Trends Biochem. Sci. 1998, 23, 461–465. [Google Scholar] [CrossRef]
- Ferrell, J.E., Jr.; Xiong, W. Bistability in Cell Signaling: How to Make Continuous Processes Discontinuous, and Reversible Processes Irreversible. Chaos 2001, 11, 227–236. [Google Scholar] [CrossRef]
- Sel’kov, Y.Y.; Nazarenko, V.G. Analysis of the Simple Open Biochemical Reaction →S→EP→ Interacting with an Enzyme-Forming System. Biophysics 1980, 25, 1031–1036. [Google Scholar]
- Nazarenko, V.G.; Reich, J.G. Theoretical Study of Oscillatory and Resonance Phenomena in an Open System with Induction of Enzyme by Substrate. Biomed. Biochim. Acta 1984, 43, 821–828. [Google Scholar]
- Segel, L.A. Simplification and Scaling. SIAM Rev. 1972, 14, 547–571. [Google Scholar] [CrossRef]
- Segel, L.A.; Slemrod, M. The Quasi-Steady-State Assumption: A Case Study In Perturbation. SIAM Rev. 1989, 31, 446–477. [Google Scholar] [CrossRef]
- Astarita, G. Dimensional Analysis, Scaling, and Orders of Magnitude. Chem. Eng. Sci. 1997, 52, 4681–4698. [Google Scholar] [CrossRef]
- Taniguchi, Y.; Choi, P.J.; Li, G.W.; Chen, H.; Babu, M.; Hearn, J.; Emili, A.; Xie, X.S. Quantifying E. coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells. Science 2010, 329, 533–538. [Google Scholar] [CrossRef] [Green Version]
- Schwanhausser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global Quantification of Mammalian Gene Expression Control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Kopell, N. Invariant Manifolds and the Initialization Problem for Some Atmospheric Equations. Physica D 1985, 14, 203–215. [Google Scholar] [CrossRef]
- Yannacopoulos, A.N.; Tomlin, A.S.; Brindley, J.; Merkin, J.H.; Pilling, M.J. The Use of Algebraic Sets in the Approximation of Inertial Manifolds and Lumping in Chemical Kinetic Systems. Physica D 1995, 83, 421–449. [Google Scholar] [CrossRef]
- Kaper, H.G.; Kaper, T.J. Asymptotic Analysis of Two Reduction Methods for Systems of Chemical Reactions. Physica D 2002, 165, 66–93. [Google Scholar] [CrossRef] [Green Version]
- Zagaris, A.; Kaper, H.G.; Kaper, T.J. Analysis of the Computational Singular Perturbation Reduction Method for Chemical Kinetics. J. Nonlinear Sci. 2004, 14, 59–91. [Google Scholar] [CrossRef] [Green Version]
- Ginoux, J.M.; Rossetto, B.; Chua, L.O. Slow Invariant Manifolds as Curvature of the Flow of Dynamical Systems. Int. J. Bifurc. Chaos 2008, 18, 3409–3430. [Google Scholar] [CrossRef] [Green Version]
- Goldfarb, I.; Gol’dshtein, V.; Maas, U. Comparative Analysis of Two Asymptotic Approaches Based on Integral Manifolds. IMA J. Appl. Math. 2004, 69, 353–374. [Google Scholar] [CrossRef]
- Van Kampen, N.G. Elimination of Fast Variables. Phys. Rep. 1985, 124, 69–160. [Google Scholar] [CrossRef]
- Tikhonov, A.N. Systems of Differential Equations Containing Small Parameters in the Derivatives. Matematicheskii Sbornik 1952, 31, 575–586. [Google Scholar]
- Tikhonov, A.N.; Vasil’eva, A.B.; Sveshnikov, A.G. Differential Equations; Springer: Berlin, Germany, 1985. [Google Scholar]
- Desroches, M.; Guckenheimer, J.; Krauskopf, B.; Kuehn, C.; Osinga, H.M.; Wechselberger, M. Mixed-Mode Oscillations with Multiple Time Scales. SIAM Rev. 2012, 54, 211–288. [Google Scholar] [CrossRef]
- Brøns, M. An Iterative Method for the Canard Explosion in General Planar Systems. Discrete Contin. Dyn. Syst. Suppl. 2013, 77–83. [Google Scholar] [CrossRef] [Green Version]
- Noethen, L.; Walcher, S. Tikhonov’s Theorem and Quasi-Steady State. Discrete Contin. Dyn. Syst. B 2011, 16, 945–961. [Google Scholar] [CrossRef]
- Nguyen, A.H.; Fraser, S.J. Geometrical picture of reaction in enzyme kinetics. J. Chem. Phys. 1989, 91, 186–193. [Google Scholar] [CrossRef]
- Skodje, R.T.; Davis, M.J. Geometrical Simplification of Complex Kinetic Systems. J. Phys. Chem. A 2001, 105, 10356–10365. [Google Scholar] [CrossRef]
- Kuehn, C. Multiple Time Scale Dynamics; Applied Mathematical Sciences; Springer: Cham, Switzerland, 2015; Volume 191. [Google Scholar] [CrossRef]
- Roberts, A.J. Low-Dimensional Modelling of Dynamics Via Computer Algebra. Comput. Phys. Commun. 1997, 100, 215–230. [Google Scholar] [CrossRef] [Green Version]
- Gorban, A.N.; Karlin, I.V.; Zmievskii, V.B.; Dymova, S.V. Reduced Description in the Reaction Kinetics. Physica A 2000, 275, 361–379. [Google Scholar] [CrossRef] [Green Version]
- Kristiansen, K.U.; Brøns, M.; Starke, J. An Iterative Method for the Approximation of Fibers in Slow-Fast Systems. SIAM J. Appl. Dyn. Syst. 2014, 13, 861–900. [Google Scholar] [CrossRef] [Green Version]
- Okeke, B.E.; Roussel, M.R. An Invariant-Manifold Approach to Lumping. Math. Model. Nat. Phenom. 2015, 10, 149–167. [Google Scholar] [CrossRef]
- Chow, S.N.; Mallet-Paret, J. Singularly Perturbed Delay-Differential Equations. In Coupled Nonlinear Oscillators; Chandra, J., Scott, A., Eds.; North-Holland: Amsterdam, The Netherlands, 1983; pp. 7–12. [Google Scholar]
- Tian, H. Asymptotic Expansion for the Solution of Singularly Perturbed Delay Differential Equations. J. Math. Anal. Appl. 2003, 281, 678–696. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Han, Q.Q.; Zhou, M.T.; Chen, X.; Guo, L. Protein Turnover Analysis in Salmonella Typhimurium during Infection by Dynamic SILAC, Topograph, and Quantitative Proteomics. J. Basic Microbiol. 2016, 56, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Viil, J.; Ivanova, H.; Pärnik, T. Estimation of Rate Constants of the Partial Reactions of Carboxylation of Ribulose-1,5-Bisphosphate In Vivo. Photosynth. Res. 1999, 60, 247–256. [Google Scholar] [CrossRef]
- McNevin, D.; von Caemmerer, S.; Farquhar, G. Determining RuBisCO Activation Kinetics and Other Rate and Equilibrium Constants by Simultaneous Multiple Non-Linear Regression of a Kinetic Model. J. Exp. Bot. 2006, 57, 3883–3900. [Google Scholar] [CrossRef] [Green Version]
- Albe, K.R.; Butler, M.H.; Wright, B.E. Cellular Concentrations of Enzymes and Their Substrates. J. Theor. Biol. 1990, 143, 163–195. [Google Scholar] [CrossRef]
- Murray, J.D. Asymptotic Analysis; Springer: New York, NY, USA, 1984. [Google Scholar]
- Xu, J.; Jiang, S. Delay-Induced Bogdanov-Takens Bifurcation and Dynamical Classifications in a Slow-Fast Flexible Joint System. Int. J. Bifurc. Chaos 2015, 25. [Google Scholar] [CrossRef]
- Maas, U. Efficient Calculation of Intrinsic Low-Dimensional Manifolds for the Simplification of Chemical Kinetics. Comput. Vis. Sci. 1998, 1, 69–81. [Google Scholar] [CrossRef]
- Gorban, A.N.; Karlin, I.V.; Zinovyev, A.Y. Invariant Grids for Reaction Kinetics. Physica A 2004, 333, 106–154. [Google Scholar] [CrossRef] [Green Version]
- Broer, H.W.; Hagen, A.; Vegter, G. Numerical Continuation of Normally Hyperbolic Invariant Manifolds. Nonlinearity 2007, 20, 1499–1534. [Google Scholar] [CrossRef]
- Roussel, M.R.; Zhu, R. Validation of an Algorithm for Delay Stochastic Simulation of Transcription and Translation in Prokaryotic Gene Expression. Phys. Biol. 2006, 3, 274–284. [Google Scholar] [CrossRef]
- Swinburne, I.A.; Silver, P.A. Intron Delays and Transcriptional Timing during Development. Dev. Cell 2008, 14, 324–330. [Google Scholar] [CrossRef] [Green Version]
- Xiao, M.; Zheng, W.X.; Cao, J. Stability and Bifurcation of Genetic Regulatory Networks with Small RNAs and Multiple Delays. Int. J. Comput. Math. 2014, 91, 907–927. [Google Scholar] [CrossRef]
- Trofimenkoff, E.A.M.; Roussel, M.R. Small Binding-Site Clearance Delays Are Not Negligible in Gene Expression Modeling. Math. Biosci. 2020, 325. [Google Scholar] [CrossRef] [PubMed]










| k | 2 | 3 | |
|---|---|---|---|
| 1 | 152 | 628 | 4.07 k |
| 2 | 4.15 k | 26.9 k | 190 k |
| 3 | 252 k | 2.09 M | 14.3 M |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roussel, M.R. Perturbative-Iterative Computation of Inertial Manifolds of Systems of Delay-Differential Equations with Small Delays. Algorithms 2020, 13, 209. https://doi.org/10.3390/a13090209
Roussel MR. Perturbative-Iterative Computation of Inertial Manifolds of Systems of Delay-Differential Equations with Small Delays. Algorithms. 2020; 13(9):209. https://doi.org/10.3390/a13090209
Chicago/Turabian StyleRoussel, Marc R. 2020. "Perturbative-Iterative Computation of Inertial Manifolds of Systems of Delay-Differential Equations with Small Delays" Algorithms 13, no. 9: 209. https://doi.org/10.3390/a13090209