Adsorption of Chiral [5]-Aza[5]helicenes on DNA Can Modify Its Hydrophilicity and Affect Its Chiral Architecture: A Molecular Dynamics Study
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Self Aggregation of 5-Aza[5]helicene Molecules at Low and at High Concentration of Enantiopure Compounds and Racemic Mixtures
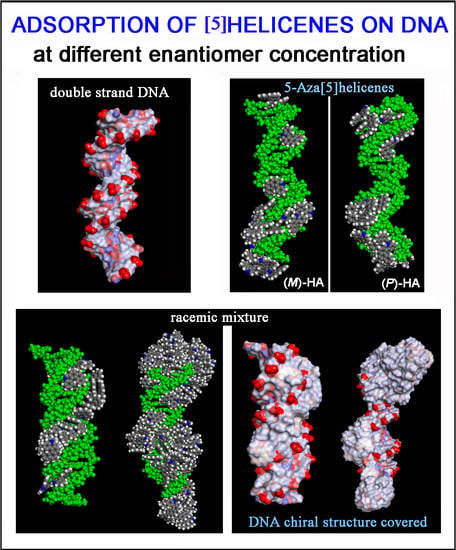
3.2. DNA/Chiral 5-Aza[5]helicene Interaction in 1:1 Stoichiometry: Importance of Chirality
3.3. DNA/5-Aza[5]helicenes Interaction: Importance of Concentration
3.3.1. DNA/Chiral 5-Aza[5]helicenes Interaction at Low Concentration
3.3.2. DNA/Chiral 5-Aza[5]helicenes Interaction at High Concentration
3.4. DNA/Chiral 5-Aza[5]helicenes Interaction in Racemic Mixtures at Two Different Concentrations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| B-DNA | right-handed double-helical structure of DNA |
| Z-DNA | a different left-handed double-helical structure of DNA |
| MM | Molecular Mechanics |
| MD | Molecular Dynamics |
| NVT | Number of particles, volume and temperature constant during the MD run |
| NP | nanoparticle |
| (M)-HA | (M)-5-Aza[5]helicene |
| (P)-HA | (P)-5-Aza[5]helicene |
| PBC | Periodic boundary conditions |
| LD | linear dichroism |
| CD | circular dichroism |
References
- Mitsui, Y.; Langridge, R.; Shortle, B.E.; Cantor, C.R.; Grant, R.C.; Kodama, M.; Wells, R.D. Physical and Enzymatic Studies on Poly d(I–C).Poly d(I–C), an Unusual Double-helical DNA. Nat. Cell Biol. 1970, 228, 1166–1169. [Google Scholar] [CrossRef] [PubMed]
- Herman, D.M.; Baird, E.E.; Dervan, P.B. Stereochemical Control of the DNA Binding Affinity, Sequence Specificity, and Orientation Preference of Chiral Hairpin Polyamides in the Minor Groove. J. Am. Chem. Soc. 1998, 120, 1382–1391. [Google Scholar] [CrossRef] [Green Version]
- Ehley, J.A.; Melander, C.; Herman, D.; Baird, E.E.; Ferguson, H.A.; Goodrich, J.A.; Dervan, P.B.; Gottesfeld, J.M. Promoter Scanning for Transcription Inhibition with DNA-Binding Polyamides. Mol. Cell. Biol. 2002, 22, 1723–1733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman, D.M.; Baird, E.E.; Dervan, P.B. Tandem Hairpin Motif for Recognition in the Minor Groove of DNA by Pyrrole-Imidazole Polyamides. Chem. A Eur. J. 1999, 5, 975–983. [Google Scholar] [CrossRef]
- Honzawa, S.; Okubo, H.; Anzai, S.; Yamaguchi, M.; Tsumoto, K.; Kumagai, I. Chiral recognition in the binding of helicenediamine to double strand DNA: Interactions between low molecular weight helical compounds and a helical polymer. Bioorg. Med. Chem. 2002, 10, 3213–3218. [Google Scholar] [CrossRef]
- Staab, H.A.; Zirnstein, M.A.; Krieger, C. New Proton Sponges 9. Benzo[1,2-h-4,3-h’]diquinoline (1,14-diaza[5]Helicene)—Synthesis, Structure, and Properties. Angew. Chem. Int. Ed. Engl. 1989, 59, 2–19. [Google Scholar] [CrossRef]
- Gauthier, E.S.; Rodriguez, R.; Crassous, J. Metal-Based Multihelicenic Architectures. Angew. Chem. Int. Ed. 2020, 58, 5562–5566. [Google Scholar] [CrossRef]
- Knöller, J.A.; Meng, G.; Wang, X.; Hall, D.; Pershin, A.; Beljonne, D.; Olivier, Y.; Laschat, S.; Zysman-Colman, E.; Wang, S. Intramolecular Borylation via Sequential B−Mes Bond Cleavage for the Divergent Synthesis of B,N,B-Doped Benzo[4]helicenes. Angew. Chem. Int. Ed. 2020, 59, 3156–3160. [Google Scholar] [CrossRef]
- Wigglesworth, T.J.; Sud, D.; Norsten, T.B.; Lekhi, V.S.; Branda, N.R. Chiral Discrimination in Photochromic Helicenes. J. Am. Chem. Soc. 2005, 127, 7272–7273. [Google Scholar] [CrossRef]
- Schulte, T.R.; Holstein, J.J.; Clever, G.H. Chiral Self-Discrimination and Guest Recognition in Helicene-Based Coordination Cages. Angew. Chem. Int. Ed. 2019, 58, 5562–5566. [Google Scholar] [CrossRef]
- Zhu, Y.; Xia, Z.; Cai, Z.; Yuan, Z.; Jiang, N.; Li, T.; Wang, Y.; Guo, X.; Li, Z.; Ma, S.; et al. Synthesis and Characterization of Hexapole [7]Helicene, A Circularly Twisted Chiral Nanographene. J. Am. Chem. Soc. 2018, 140, 4222–4226. [Google Scholar] [CrossRef]
- Gingras, M. One hundred years of helicene chemistry. Part 3: Applications and properties of carbohelicenes. Chem. Soc. Rev. 2013, 42, 1051–1095. [Google Scholar] [CrossRef]
- Li, M.; Yao, W.; Chen, J.-D.; Lu, H.-Y.; Zhao, Y.; Chen, C.-F. Tetrahydro[5]helicene-based full-color emission dyes in both solution and solid states: Synthesis, structures, photophysical properties and optical waveguide applications. J. Mater. Chem. C 2014, 2, 8373–8380. [Google Scholar] [CrossRef]
- Gauthier, E.S.; Abella, L.; Hellou, N.; Darquie, B.; Caytan, E.; Roisnel, T.; Vanthuyne, N.; Favereau, L.; Srebro-Hooper, M.; Williams, J.A.G.; et al. Long-Lived Circularly Polarized Phosphorescence in Helicene-NHC Rhenium(I) Complexes: The Influence of Helicene, Halogen, and Stereochemistry on Emission Properties. Angew. Chem. Int. Ed. 2020, 59, 8394–8400. [Google Scholar] [CrossRef]
- Dhbaibi, K.; Favereau, L.; Srebro-Hooper, M.; Quinton, C.; Vanthuyne, N.; Arrico, L.; Roisnel, T.; Jamoussi, B.; Poriel, C.; Cabanetos, C.; et al. Modulation of circularly polarized luminescence through excited-state symmetry breaking and interbranched exciton coupling in helical push–pull organic systems. Chem. Sci. 2020, 11, 567–576. [Google Scholar] [CrossRef] [Green Version]
- Otani, T.; Sasayama, T.; Iwashimizu, C.; Kanyiva, K.S.; Kawai, H.; Shibata, T. Short-step synthesis and chiroptical properties of polyaza[5]–[9]helicenes with blue to green-colour emission. Chem. Commun. 2020, 56, 4484–4487. [Google Scholar] [CrossRef]
- Shen, C.; Gan, F.; Zhang, G.; Ding, Y.; Wang, J.; Wang, R.; Crassous, J.; Qiu, H. Helicene-derived aggregation-induced emission conjugates with highly tunable circularly polarized luminescence. Mater. Chem. Front. 2020, 4, 837–844. [Google Scholar] [CrossRef]
- Saleh, N.; Shen, C.; Crassous, J. Helicene-based transition metal complexes: Synthesis, properties and applications. Chem. Sci. 2014, 5, 3680–3694. [Google Scholar] [CrossRef]
- Passeri, R.; Aloisi, G.G.; Elisei, F.; Latterini, L.; Caronna, T.; Fontana, F.; Sora, I.N. Photophysical properties of N-alkylated azahelicene derivatives as DNA intercalators: Counterion effects. Photochem. Photobiol. Sci. 2009, 8, 1574–1582. [Google Scholar] [CrossRef]
- Lebon, F.; Longhi, G.; Gangemi, F.; Abbate, S.; Priess, J.; Juza, M.; Bazzini, C.; Caronna, T.; Mele, A. Chiroptical Properties of Some Monoazapentahelicenes. J. Phys. Chem. A 2004, 108, 11752–11761. [Google Scholar] [CrossRef]
- Kel, O.; Fürstenberg, A.; Mehanna, N.; Nicolas, C.; Laleu, B.; Hammarson, M.; Albinsson, B.; Lacour, J.; Vauthey, E. Chiral Selectivity in the Binding of [4]Helicene Derivatives to Double-Stranded DNA. Chem. A Eur. J. 2013, 19, 7173–7180. [Google Scholar] [CrossRef]
- Tsuji, G.; Kawakami, K.; Sasaki, S. Enantioselective binding of chiral 1,14-dimethyl[5]helicene–spermine ligands with B- and Z-DNA. Bioorg. Med. Chem. 2013, 21, 6063–6068. [Google Scholar] [CrossRef] [Green Version]
- Kawara, K.; Tsuji, G.; Taniguchi, Y.; Sasaki, S. Synchronized Chiral Induction between [5]Helicene-Spermine Ligand andB-ZDNA Transition. Chem. A Eur. J. 2017, 23, 1763–1769. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, Y.X.; Sugiyama, H.; Umano, T.; Osuga, H.; Tanaka, K. (P)-Helicene Displays Chiral Selection in Binding to Z-DNA. J. Am. Chem. Soc. 2004, 126, 6566–6567. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, C.; Brovelli, S.; Caronna, T.; Gambarotti, C.; Giannone, M.; Macchi, P.; Meinardi, F.; Mele, A.; Panzeri, W.; Recupero, F.; et al. Synthesis and Characterization of Some Aza[5]helicenes. Eur. J. Org. Chem. 2005, 2005, 1247–1257. [Google Scholar] [CrossRef]
- Abbate, S.; Caronna, T.; Longo, A.; Ruggirello, A.; Liveri, V.T. Study of Confined 5-Aza[5]helicene in Ytterbium(III) Bis(2-ethylhexyl) Sulfosuccinate Reversed Micelles. J. Phys. Chem. B 2007, 111, 4089–4097. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Protein Adsorption on Biomaterial and Nanomaterial Surfaces: A Molecular Modeling Approach to Study Non-Covalent Interactions. J. Appl. Biomater. Biomech. 2010, 8, 135–145. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Molecular dynamics study of host–guest interactions in cyclodextrins: Methodology and data analysis for a comparison with solution data and the solid-state structure. J. Incl. Phenom. Macrocycl. Chem. 2007, 57, 683–688. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Adsorption of charged albumin subdomains on a graphite surface. J. Biomed. Mater. Res. A 2006, 76, 638–645. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Surface Topography Effects in Protein Adsorption on Nanostructured Carbon Allotropes. Langmuir 2013, 29, 4883–4893. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Separation of chiral nanotubes with an opposite handedness by chiral oligopeptide adsorption: A molecular dynamics study. J. Chromatogr. A 2015, 1425, 221–230. [Google Scholar] [CrossRef]
- Raffaini, G.; Melone, L.; Punta, C. Understanding the topography effects on competitive adsorption on a nanosized anatase crystal: A molecular dynamics study. Chem. Commun. 2013, 49, 7581. [Google Scholar] [CrossRef]
- Raffaini, G.; Mazzaglia, A.; Ganazzoli, F. Aggregation behaviour of amphiphilic cyclodextrins: The nucleation stage by atomistic molecular dynamics simulations. Beilstein J. Org. Chem. 2015, 11, 2459–2473. [Google Scholar] [CrossRef] [Green Version]
- Raffaini, G.; Ganazzoli, F. Understanding surface interaction and inclusion complexes between piroxicam and native or crosslinked β-Cyclodextrins: The role of the drug concentration. Molecules 2020, 24, 2848. [Google Scholar] [CrossRef]
- Dauber-Osguthorpe, P.; Roberts, V.A.; Osguthorpe, D.J.; Wolff, J.; Genest, M.; Hagler, A.T. Structure and Energetics of Ligand-Binding to Proteins—Escherichia-Coli Dihydrofolate Reductase Trimethoprim Drug-Receptor System. Proteins 1988, 4, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Hwang, M.-J.; Ni, X.; Waldman, M.; Ewig, C.S.; Hagler, A.T. Derivation of class II force fields. VI. Carbohydrate compounds and anomeric effects. Biopolymers 1998, 45, 435–468. [Google Scholar] [CrossRef]
- Aminpour, M.; Montemagno, C.; Tuszynski, J.A. An Overview of Molecular Modeling for Drug Discovery with Specific Illustrative Examples of Applications. Molecules 2019, 24, 1693. [Google Scholar] [CrossRef] [Green Version]
- Dubbeldam, D.; Walton, K.S.; Vlugt, T.J.H.; Calero, S. Design, Parameterization, and Implementation of Atomic Force Fields for Adsorption in Nanoporous Materials. Adv. Theory Simul. 2019, 2, 1900135–1900197. [Google Scholar] [CrossRef]
- Bestel, I.; Campins, N.; Marchenko, A.; Fichou, D.; Grinstaff, M.W.; Barthélémy, P. Two-dimensional self-assembly and complementary base-pairing between amphiphile nucleotides on graphite. J. Colloid Interface Sci. 2008, 323, 435–440. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, M.L.; Cranford, S.W. Ranking of Molecular Biomarker Interaction with Targeted DNA Nucleobases via Full Atomistic Molecular Dynamics. Sci. Rep. 2016, 6, 18659. [Google Scholar] [CrossRef] [Green Version]
- Materials Studio BIOVIA. Accelrys Inc. InsightII 2000; Accelrys Inc.: San Diego, CA, USA, 2000; Available online: https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-materials-studio/ (accessed on 10 September 2020).
- Mondragón, A.; Harrison, S. The phage 434 complex at 2.5 A resolution. J. Mol. Biol. 1991, 219, 321–334. [Google Scholar] [CrossRef]
- Ichimura, K. Photoalignment of Liquid-Crystal Systems. Chem. Rev. 2000, 100, 1847–1874. [Google Scholar] [CrossRef]
- Sergeyev, S.; Pisula, W.; Geerts, Y.H. Discotic liquid crystals: A new generation of organic semiconductors. Chem. Soc. Rev. 2007, 36, 1902–1929. [Google Scholar] [CrossRef]
- Laschat, S.; Baro, A.; Steinke, N.; Giesselmann, F.; Hagele, C.; Scalia, G.; Judele, R.; Kapatsina, E.; Sauer, S.; Schreivogel, A.; et al. Discotic liquid crystals: From tailor-made synthesis to plastic electronics. Angew. Chem. Int. Ed. 2007, 46, 4832–4887. [Google Scholar] [CrossRef]
- Mitov, M. Cholesteric Liquid Crystals with a Broad Light Reflection Band. Adv. Mater. 2012, 24, 6260–6276. [Google Scholar] [CrossRef] [PubMed]
- Tamaoki, N. Cholesteric Liquid Crystals for Color Information Technology. Adv. Mater. 2001, 13, 1135–1147. [Google Scholar] [CrossRef]
- Latterini, L.; Galletti, E.; Passeri, R.; Barbafina, A.; Urbanelli, L.; Emiliani, C.; Elisei, F.; Fontana, F.; Mele, A.; Caronna, T. Fluorescence properties of aza-helicenium derivatives for cell imaging. J. Photochem. Photobiol. A Chem. 2011, 222, 307–313. [Google Scholar] [CrossRef]
- Cheriya, R.T.; Joy, J.; Rajagopal, S.K.; Nagarajan, K.; Hariharan, M. DNA-Enforced Conformational Restriction of an Atropisomer. J. Phys. Chem. C 2012, 116, 22631–22636. [Google Scholar] [CrossRef]










| Site of Interaction | (M)-HA Eint (kJ/mol) | (P)-HA Eint (kJ/mol) | Θ Value (°) |
|---|---|---|---|
| Minor groove | −176.4 | −181.0 | −31.055° |
| Major groove | −166.0 | −186.1 | 30.887° |
| (M)-HA Θ Value (°) | (P)-HA Θ Value (°) | Racemic Mixture Θ Value (°) |
|---|---|---|
| −34.845 | 28.906 | −33.509 |
| −34.042 | 32.411 | 29.600 |
| −34.097 | 30.561 | 29.335 |
| −34.309 | 30.143 | −35.281 |
| −34.571 | 29.894 | −32.191 |
| −29.006 | 32.999 | −29.811 |
| −30.403 | 30.907 | −28.395 |
| −35.449 | 33.004 | −31.265 |
| −27.265 | 33.139 | 27.465 |
| −35.485 | 35.388 | 30.308 |
| −29.408 | 30.426 | 34.277 |
| −31.562 | 29.240 | 30.720 |
| −32.144 | 29.051 | −32.220 |
| −31.986 | 32.635 | −31.172 |
| −32.355 | 31.983 | −29.936 |
| −32.067 | 29.356 | 31.234 |
| −29.843 | 29.356 | 29.065 |
| −31.724 | 34.295 | 30.204 |
| −28.727 | 32.277 | −33.123 |
| −28.275 | 30.206 | 32.905 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raffaini, G.; Mele, A.; Caronna, T. Adsorption of Chiral [5]-Aza[5]helicenes on DNA Can Modify Its Hydrophilicity and Affect Its Chiral Architecture: A Molecular Dynamics Study. Materials 2020, 13, 5031. https://doi.org/10.3390/ma13215031
Raffaini G, Mele A, Caronna T. Adsorption of Chiral [5]-Aza[5]helicenes on DNA Can Modify Its Hydrophilicity and Affect Its Chiral Architecture: A Molecular Dynamics Study. Materials. 2020; 13(21):5031. https://doi.org/10.3390/ma13215031
Chicago/Turabian StyleRaffaini, Giuseppina, Andrea Mele, and Tullio Caronna. 2020. "Adsorption of Chiral [5]-Aza[5]helicenes on DNA Can Modify Its Hydrophilicity and Affect Its Chiral Architecture: A Molecular Dynamics Study" Materials 13, no. 21: 5031. https://doi.org/10.3390/ma13215031