Exploring the Potential of Crassostrea nippona Hydrolysates as Dietary Supplements for Mitigating Dexamethasone-Induced Muscle Atrophy in C2C12 Cells
Abstract
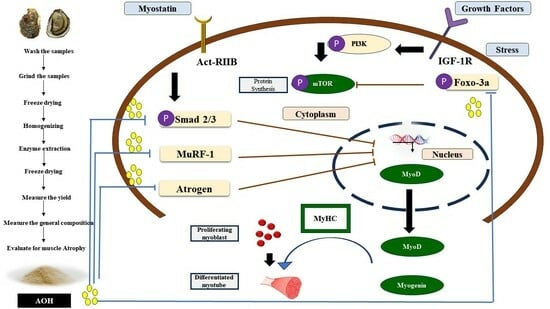
:1. Introduction
2. Results
2.1. General Composition of the Enzyme Hydrolysates of Crassostrea Nippona
2.2. Cell Cytotoxicity and Proliferation Activity of the Enzyme Hydrolysates of Crassostrea Nippona
2.3. Immunostaining and Myotube Diameters on MyHC Expression
2.4. Molecular Weight Determination and Amino Acid Composition
2.5. Protein Expression after Treatment with AOH
2.6. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of Enzyme Hydrolysate of Crassostrea Nippona
4.3. General Composition of Crassostrea Nippona
4.4. Cell Culture and Cell Differentiation
4.5. Cytotoxicity Evaluation using Cell Counting Kit-8 (CCK-8 assay)
4.6. Cell Proliferation Evaluation using 5-Bromo-2ʹ-deoxyuridine (BrdU) Assay
4.7. Immunostaining of Myosin Heavy Chain (MyHC)
4.8. Molecular Weight Determination and Amino Acid Composition
4.9. Western Blot Analysis
4.10. RT-qPCR
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Jackman, R.W.; Kandarian, S.C. The molecular basis of skeletal muscle atrophy. Am. J. Physiol. Cell Physiol. 2004, 287, C834–C843. [Google Scholar] [CrossRef] [PubMed]
- Morley, J.E.; Anker, S.D.; von Haehling, S. Prevalence, incidence, and clinical impact of sarcopenia: Facts, numbers, and epidemiology—Update 2014. J. Cachexia Sarcopenia Muscle 2014, 5, 253–259. [Google Scholar] [CrossRef]
- Tipton, K.D.; Elliott, T.A.; Cree, M.G.; Aarsland, A.A.; Sanford, A.P.; Wolfe, R.R. Stimulation of net muscle protein synthesis by whey proteingestion before and after exercise. Am. J. Physiol. Endocrinol. Metab. 2007, 292, E71–E76. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Dyar, K.A.; Ciciliot, S.; Blaauw, B.; Sandri, M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013, 280, 4294–4314. [Google Scholar] [CrossRef] [PubMed]
- Sartori, R.; Milan, G.; Patron, M.; Mammucari, C.; Blaauw, B.; Abraham, R.; Sandri, M. Smad2 and 3 transcription factors control muscle mass in adulthood. Am. J. Physiol. Cell Physiol. 2009, 296, C1248–C1257. [Google Scholar] [CrossRef] [PubMed]
- Nagahawatta, D.P.; Liyanage, N.M.; Jayawardena, T.U.; Jayawardhana, H.H.A.C.K.; Oh, J.-Y.; Sanjeewa, K.K.A.; Kang, S.I.; Jeon, Y.-J. Skeletal muscle growth activity of Olive Flounder (Paralichthys olivaceus) meat digest. Food Biosci. 2023, 53, 102809. [Google Scholar] [CrossRef]
- Jodral-Segado, A.M.; Navarro-Alarcón, M.; López-G de la Serrana, H.; López-Martínez, M.a.C. Magnesium and calcium contents in foods from SE Spain: Influencing factors and estimation of daily dietary intakes. Sci. Total Environ. 2003, 312, 47–58. [Google Scholar] [CrossRef]
- Hao, L.; Wang, X.; Cao, Y.; Xu, J.; Xue, C. A comprehensive review of oyster peptides: Preparation, characterisation and bioactivities. Rev. Aquac. 2022, 14, 120–138. [Google Scholar] [CrossRef]
- Rajapakse, N.; Mendis, E.; Jung, W.-K.; Je, J.-Y.; Kim, S.-K. Purification of a radical scavenging peptide from fermented mussel sauce and its antioxidant properties. Food Res. Int. 2005, 38, 175–182. [Google Scholar] [CrossRef]
- Jung, W.-K.; Kim, S.-K. Isolation and characterisation of an anticoagulant oligopeptide from blue mussel, Mytilus edulis. Food Chem. 2009, 117, 687–692. [Google Scholar] [CrossRef]
- Anderson, R.S.; Beaven, A.E. Antibacterial activities of oyster (Crassostrea virginica) and mussel (Mytilus edulis and Geukensia demissa) plasma. Aquat. Living Resour. 2001, 14, 343–349. [Google Scholar] [CrossRef]
- Imsong, L.; Murali, M. DNA barcoding, determination of bioactive compounds, Antioxidant and anti-diabetic property in edible gastropod Brotia costula (Rafinesque, 1833) of dimapur district, Nagaland. Int. J. Pharm. Sci. Res. 2022, 14, 1795–1804. [Google Scholar]
- Hu, X.; Tao, N.; Wang, X.; Xiao, J.; Wang, M. Marine-derived bioactive compounds with anti-obesity effect: A review. J. Funct. Foods. 2016, 21, 372–387. [Google Scholar] [CrossRef]
- La Paglia, L.; Vazzana, M.; Mauro, M.; Urso, A.; Arizza, V.; Vizzini, A. Bioactive Molecules from the Innate Immunity of Ascidians and Innovative Methods of Drug Discovery: A Computational Approach Based on Artificial Intelligence. Mar. Drugs 2024, 22, 6. [Google Scholar] [CrossRef]
- Librizzi, M.; Martino, C.; Mauro, M.; Abruscato, G.; Arizza, V.; Vazzana, M.; Luparello, C. Natural Anticancer Peptides from Marine Animal Species: Evidence from In Vitro Cell Model Systems. Cancers 2024, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Punginelli, D.; Catania, V.; Abruscato, G.; Luparello, C.; Vazzana, M.; Mauro, M.; Cunsolo, V.; Saletti, R.; Di Francesco, A.; Arizza, V.; et al. New Bioactive Peptides from the Mediterranean Seagrass Posidonia oceanica (L.) Delile and Their Impact on Antimicrobial Activity and Apoptosis of Human Cancer Cells. Int. J. Mol. Sci. 2023, 24, 5650. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Branni, R.; Abruscato, G.; Lazzara, V.; Sugár, S.; Arizza, V.; Mauro, M.; Di Stefano, V.; Vazzana, M. Biological and Proteomic Characterization of the Anti-Cancer Potency of Aqueous Extracts from Cell-Free Coelomic Fluid of Arbacia lixula Sea Urchin an In Vitro Model of Human Hepatocellular Carcinoma. J. Mar. Sci. Eng. 2022, 10, 1292. [Google Scholar] [CrossRef]
- Punginelli, D.; Schillaci, D.; Mauro, M.; Deidun, A.; Barone, G.; Arizza, V.; Vazzana, M. The potential of antimicrobial peptides isolated from freshwater crayfish species in new drug development: A review. Dev. Comp. Immunol. 2022, 126, 104258. [Google Scholar] [CrossRef]
- Mauro, M.; Lazzara, V.; Punginelli, D.; Arizza, V.; Vazzana, M. Antitumoral compounds from vertebrate sister group: A review of Mediterranean ascidians. Dev. Comp. Immunol. 2020, 108, 103669. [Google Scholar] [CrossRef]
- Mauro, M.; Pinto, P.; Settanni, L.; Puccio, V.; Vazzana, M.; Hornsby, B.L.; Fabbrizio, A.; Di Stefano, V.; Barone, G.; Arizza, V. Chitosan Film Functionalized with Grape Seed Oil—Preliminary Evaluation of Antimicrobial Activity. Dev. Comp. Immunol. 2022, 14, 5410. [Google Scholar] [CrossRef]
- Kios, K.; Kakasis, S.; Syropoulou, F.; Boziaris, I.S. Chapter 10—Seafood and shellfish. In Functional Foods and Their Implications for Health Promotion; Zabetakis, I., Tsoupras, A., Lordan, R., Ramji, D., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 281–302. [Google Scholar]
- de Souza, M.M.M.; da Boa Morte, E.S.; Cardoso, L.G.; Nunes, D.V.; de Souza, C.O.; Druzian, J.I.; Cardoso, R.d.C.V. Nutritional contribution of shellfish from the biodiversity of Todos os Santos Bay, Brazil. J. Food Compos. Anal. 2021, 102, 103999. [Google Scholar] [CrossRef]
- Botta, R.; Asche, F.; Borsum, J.S.; Camp, E.V. A review of global oyster aquaculture production and consumption. Mar. Policy 2020, 117, 103952. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, D.; Do, K.; Yang, C.B.; Jeon, S.W.; Jang, A. Effects of Horse Meat Hydrolysate on Oxidative Stress, Proinflammatory Cytokines, and the Ubiquitin-Proteasomal System of C2C12 Cells. Food Sci. Anim. Resour. 2024, 44, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.; Kang, S.-I.; Lee, S.-W.; Amarasiri, R.P.G.S.K.; Nagahawatta, D.P.; Roh, Y.; Wang, L.; Ryu, B.; Jeon, Y.-J. Exploring the Potential of Olive Flounder Processing By-Products as a Source of Functional Ingredients for Muscle Enhancement. Antioxidants 2023, 12, 1755. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.-H.; Choung, S.-Y. Oyster Hydrolysates Attenuate Muscle Atrophy via Regulating Protein Turnover and Mitochondria Biogenesis in C2C12 Cell and Immobilized Mice. Nutrients 2021, 13, 4385. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, Q.; Xu, C.; Liu, S.; Kong, L.; Yu, H. Response to selection for growth in successive mass selected generations of Iwagaki oyster Crassostrea nippona. Aquaculture 2022, 560, 738575. [Google Scholar] [CrossRef]
- Gong, J.; Li, Q.; Yu, H.; Liu, S.; Kong, L. First de novo transcriptome assembly of Iwagaki oyster, Crassostrea nippona, and comparative evolutionary analysis of salinity-stress response genes in Crassostrea oysters. Mar. Genom. 2021, 56, 100805. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Li, Q.; Yu, H.; Liu, S.; Kong, L. Effects of low salinity on hemolymph osmolality and transcriptome of the Iwagaki oyster, Crassostrea nippona. Fish Shellfish Immunol. 2022, 126, 211–216. [Google Scholar] [CrossRef]
- Venugopal, V.; Gopakumar, K. Shellfish: Nutritive Value, Health Benefits, and Consumer Safety. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1219–1242. [Google Scholar] [CrossRef]
- Hong, Y.J.; Saroj, R.K.; Park, W.I.; Yi, G.-C. One-dimensional semiconductor nanostructures grown on two-dimensional nanomaterials for flexible device applications. APL Mater. 2021, 9, 060907. [Google Scholar] [CrossRef]
- Ko, S.-C.; Kang, N.; Kim, E.-A.; Kang, M.C.; Lee, S.-H.; Kang, S.-M.; Lee, J.-B.; Jeon, B.-T.; Kim, S.-K.; Park, S.-J.; et al. A novel angiotensin I-converting enzyme (ACE) inhibitory peptide from a marine Chlorella ellipsoidea and its antihypertensive effect in spontaneously hypertensive rats. Process. Biochem. 2012, 47, 2005–2011. [Google Scholar] [CrossRef]
- Ryan, J.T.; Ross, R.P.; Bolton, D.; Fitzgerald, G.F.; Stanton, C. Bioactive Peptides from Muscle Sources: Meat and Fish. Nutrients 2011, 3, 765–791. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Robbens, J.; Heyndrickx, M.; Debode, J.; Raes, K. Bioprocessing of marine crustacean side-streams into bioactives: A review. JCTB 2021, 96, 1465–1474. [Google Scholar] [CrossRef]
- Yoon, J.H.; Lee, S.-M.; Lee, Y.; Kim, M.J.; Yang, J.W.; Choi, J.Y.; Kwak, J.Y.; Lee, K.-P.; Yang, Y.R.; Kwon, K.-S. Alverine citrate promotes myogenic differentiation and ameliorates muscle atrophy. Biochem. Biophys. Res. Commun. 2022, 586, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Jayawardhana, H.H.A.C.K.; Liyanage, N.M.; Nagahawatta, D.P.; Lee, H.-G.; Jeon, Y.-J.; Kang, S.I. Pepsin Hydrolysate from Surimi Industry-Related Olive Flounder Head Byproducts Attenuates LPS-Induced Inflammation and Oxidative Stress in RAW 264.7 Macrophages and In Vivo Zebrafish Model. Mar. Drugs 2024, 22, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Kuo, C.-H.; Lee, C.-L.; Kuo, W.-C.; Tsai, M.-L.; Sun, P.-P. Enzyme-Assisted Aqueous Extraction of Cobia Liver Oil and Protein Hydrolysates with Antioxidant Activity. Catalysts 2020, 10, 1323. [Google Scholar] [CrossRef]
- Wang, L.; Hu, J.; Lv, W.; Lu, W.; Pei, D.; Lv, Y.; Wang, W.; Zhang, M.; Ding, R.; Lv, M. Optimized extraction of astaxanthin from shrimp shells treated by biological enzyme and its separation and purification using macroporous resin. Food Chem. 2021, 363, 130369. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.C.; Ngo, K.N.; Tran, H.K.; Barrow, C.J. Enzyme-Assisted Coextraction of Phenolics and Polysaccharides from Padina gymnospora. Mar. Drugs 2024, 22, 42. [Google Scholar] [CrossRef]
- Ngo, D.-H.; Vo, T.-S.; Ngo, D.-N.; Wijesekara, I.; Kim, S.-K. Biological activities and potential health benefits of bioactive peptides derived from marine organisms. Int. J. Biol. Macromol. 2012, 51, 378–383. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, L.; Li, S. Advances in nutritional supplementation for sarcopenia management. Front. Nutr. 2023, 10, 1189522. [Google Scholar] [CrossRef]
- Nagahawatta, D.; Sanjeewa, K.; Jayawardena, T.U.; Kim, H.-S.; Yang, H.-W.; Jiang, Y.; Je, J.-G.; Lee, T.-K.; Jeon, Y.-J. Drying seaweeds using hybrid hot water Goodle dryer (HHGD): Comparison with freeze-dryer in chemical composition and antioxidant activity. Fish. Aquat. Sci. 2021, 24, 19–31. [Google Scholar] [CrossRef]
- Qiao, M.; Tu, M.; Wang, Z.; Mao, F.; Chen, H.; Qin, L.; Du, M. Identification and Antithrombotic Activity of Peptides from Blue Mussel (Mytilus edulis) Protein. Int. J. Mol. Sci. 2018, 19, 138. [Google Scholar] [CrossRef] [PubMed]
- Tavano, O.L. Protein hydrolysis using proteases: An important tool for food biotechnology. J. Mol. Catal. B Enzym. 2013, 90, 1–11. [Google Scholar] [CrossRef]
- Rodrigues, R.B.; Lui, T.A.; Neu, D.H.; Boscolo, W.R.; Bittencourt, F.J.R.C.A. Valine in diets for Nile tilapia. Rev. Cienc. Agron. 2018, 49, 467–474. [Google Scholar] [CrossRef]
- Kamei, Y.; Hatazawa, Y.; Uchitomi, R.; Yoshimura, R.; Miura, S. Regulation of Skeletal Muscle Function by Amino Acids. Nutrients 2020, 12, 261. [Google Scholar] [CrossRef] [PubMed]
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.-M.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Na, E.; Dharmarajan, K.; et al. Identification of Ubiquitin Ligases Required for Skeletal Muscle Atrophy. Science 2001, 294, 1704–1708. [Google Scholar] [CrossRef] [PubMed]
- Cid-Díaz, T.; Santos-Zas, I.; González-Sánchez, J.; Gurriarán-Rodríguez, U.; Mosteiro, C.S.; Casabiell, X.; García-Caballero, T.; Mouly, V.; Pazos, Y.; Camiña, J.P. Obestatin controls the ubiquitin–proteasome and autophagy–lysosome systems in glucocorticoid-induced muscle cell atrophy. J. Cachexia Sarcopenia Muscle 2017, 8, 974–990. [Google Scholar] [CrossRef]
- Torres-Velarde, J.; Llera-Herrera, R.; García-Gasca, T.; García-Gasca, A. Mechanisms of stress-related muscle atrophy in fish: An ex vivo approach. Mech. Dev. 2018, 154, 162–169. [Google Scholar] [CrossRef]
- Otsuka, Y.; Egawa, K.; Kanzaki, N.; Izumo, T.; Rogi, T.; Shibata, H. Quercetin glycosides prevent dexamethasone-induced muscle atrophy in mice. Biochem. Biophys. Rep. 2019, 18, 100618. [Google Scholar] [CrossRef]
- Chen, K.; Gao, P.; Li, Z.; Dai, A.; Yang, M.; Chen, S.; Su, J.; Deng, Z.; Li, L. Forkhead Box O Signaling Pathway in Skeletal Muscle Atrophy. Am. J. Pathol. 2022, 192, 1648–1657. [Google Scholar] [CrossRef]
- Brunetti, A.; Goldfine, I.D. Role of myogenin myoblast differentiation and its regulation by fibroblast growth factor. J. Biol. Chem. 1990, 265, 5960–5963. [Google Scholar] [CrossRef] [PubMed]
- Legerlotz, K.; Smith, H.K. Role of MyoD in denervated, disused, and exercised muscle. Muscle Nerve 2008, 38, 1087–1100. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Guan, K.-L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019, 21, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Wells, L.; Edwards, K.A.; Bernstein, S.I. Myosin heavy chain isoforms regulate muscle function but not myofibril assembly. EMBO J. 1996, 15, 4454–4459. [Google Scholar] [CrossRef] [PubMed]
- Marina, T.; Marija, C.; Ida, R. Functional Foods and the Young. J. Food Prod. Mark. 2014, 20, 441–451. [Google Scholar] [CrossRef]
- Smith, G.I.; Julliand, S.; Reeds, D.N.; Sinacore, D.R.; Klein, S.; Mittendorfer, B. Fish oil–derived n-3 PUFA therapy increases muscle mass and function in healthy older adults. Am. J. Clin. Nutr. 2015, 102, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.; Dominguez-López, I.; Lamuela-Raventós, R.M. The Chemistry Behind the Folin–Ciocalteu Method for the Estimation of (Poly)phenol Content in Food: Total Phenolic Intake in a Mediterranean Dietary Pattern. J. Agric. Food Chem. 2023, 71, 17543–17553. [Google Scholar] [CrossRef] [PubMed]
- Premarathna, A.D.; Ahmed, T.A.E.; Kulshreshtha, G.; Humayun, S.; Shormeh Darko, C.N.; Rjabovs, V.; Hammami, R.; Critchley, A.T.; Tuvikene, R.; Hincke, M.T. Polysaccharides from red seaweeds: Effect of extraction methods on physicochemical characteristics and antioxidant activities. Food Hydrocoll. 2024, 147, 109307. [Google Scholar] [CrossRef]
- Kimira, Y.; Osawa, K.; Osawa, Y.; Mano, H. Preventive Effects of Collagen-Derived Dipeptide Prolyl-Hydroxyproline against Dexamethasone-Induced Muscle Atrophy in Mouse C2C12 Skeletal Myotubes. Biomolecules 2023, 13, 1617. [Google Scholar] [CrossRef]
- Amarasiri, R.P.G.S.K.; Hyun, J.; Lee, S.-W.; Kim, J.; Jeon, Y.-J.; Lee, J.-S. Alcalase-Assisted Mytilus edulis Hydrolysate: A Nutritional Approach for Recovery from Muscle Atrophy. Mar. Drugs 2023, 21, 623. [Google Scholar] [CrossRef]
- Nagahawatta, D.P.; Liyanage, N.M.; Jayawardhana, H.H.A.C.K.; Lee, H.-G.; Jayawardena, T.U.; Jeon, Y.-J. Anti-Fine Dust Effect of Fucoidan Extracted from Ecklonia maxima Leaves in Macrophages via Inhibiting Inflammatory Signaling Pathways. Mar. Drugs 2022, 20, 413. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, X.; Zang, X.; Liu, B.; Arunakumara, K.K.I.U.; Xu, D.; Zhang, X. Growth, feed efficiency, body muscle composition, and histology of flounder (Paralichthys olivaceus) fed GH transgenic Synechocystis. Aquaculture 2008, 277, 78–82. [Google Scholar] [CrossRef]
- Cai, L.; Qin, X.; Xu, Z.; Song, Y.; Jiang, H.; Wu, Y.; Ruan, H.; Chen, J. Comparison of Cytotoxicity Evaluation of Anticancer Drugs between Real-Time Cell Analysis and CCK-8 Method. ACS Omega 2019, 4, 12036–12042. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.U.; Kim, H.-S.; Sanjeewa, K.K.A.; Kim, S.-Y.; Rho, J.-R.; Jee, Y.; Ahn, G.; Jeon, Y.-J. Sargassum horneri and isolated 6-hydroxy-4,4,7a-trimethyl-5,6,7,7a-tetrahydrobenzofuran-2(4H)-one (HTT); LPS-induced inflammation attenuation via suppressing NF-κB, MAPK and oxidative stress through Nrf2/HO-1 pathways in RAW 264.7 macrophages. Algal Res. 2019, 40, 101513. [Google Scholar] [CrossRef]
- Doyuk, F.; Dost, K. Simultaneous determination of six antibiotics belonging to four different classes in chicken meat BY HPLC/DAD and verification BY LC-MS/MS. Food Chem. 2023, 426, 136549. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Boiani, M.; He, T.; Wichers, H.J.; Hettinga, K.A. Heating affects protein digestion of skimmed goat milk under simulated infant conditions. Food Chem. 2023, 402, 134261. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-W.; Meng, J.; Cui, J.; Luan, Y.-S. Characterization and Function of MicroRNA∗s in Plants. Front. Plant Sci. 2017, 8, 2200. [Google Scholar] [CrossRef]






| Component | Composition | |||
|---|---|---|---|---|
| DWO | AOH | FOH | POH | |
| Polysaccharide | 2.72 ± 0.47% | 23.83 ± 1.42% **** | 6.86 ± 0.45% **** | 11.83 ± 0.50% **** |
| Polyphenol | 0.47 ± 0.01% | 0.51 ± 0.00% **** | 0.45 ± 0.01% | 0.50 ± 0.01% ** |
| Proteins | 40.05 ± 0.43% | 45.90 ± 2.71% **** | 36.02 ± 2.42% | 43.68 ± 1.57% **** |
| Lipids | 6.54 ± 1.56% | 8.94 ± 1.67% **** | 8.14 ± 1.42% **** | 8.50 ± 1.22% **** |
| Ash | 14.00 ± 1.00% | 75.33 ± 1.08% **** | 68.33 ± 0.58% **** | 61.33 ± 0.58% **** |
| Yield | 0.47 ± 0.01% | 0.51 ± 0.00% **** | 0.45 ± 0.01% | 0.50 ± 0.01% ** |
| Amino Acid | Moles Percentage (%) | g/100 g |
|---|---|---|
| Cysteine and cystine | 0.39% | 0.223 |
| Asparagine and asparatic acid | 8.36% | 3.429 |
| Glutamine and glutamic acid | 11.02% | 4.997 |
| Serine | 4.62% | 1.503 |
| Glycine | 10.21% | 2.361 |
| Histidine | 1.50% | 0.715 |
| Arginine | 15.43% | 8.284 |
| Threonine | 4.66% | 1.709 |
| Alanine | 9.70% | 2.665 |
| Proline | 4.17% | 1.478 |
| Tyrosine | 1.87% | 1.046 |
| Valine | 5.46% | 1.972 |
| Methionine | 2.19% | 1.008 |
| Isoleucine | 4.65% | 1.881 |
| Leucine | 6.95% | 2.809 |
| Phenylalanine | 3.06% | 1.558 |
| Tryptophan | 0.00% | 0.000 |
| Lysine | 6.12% | 2.758 |
| Total | 100.00% | 40.173 |
| Gene | Primer | Sequence |
|---|---|---|
| GAPDH | Sense Antisense | 5′-AAGGGTCATCATCTCTGCCC-3′ 5′-CCACGATGGACGTAAGGGAG-3′ |
| MyoD | Sense Antisense | 5′-GCCGCCTGAGCAAAGTGAATG-3′ 5′-CAGCGGTCCAGTGCGTAGAAG3′ |
| Myogenin | Sense Antisense | 5′-GTCCCAACCCAGGAGATCAT-3′ 5′-CCACGATGGACGTAAGGGAG-3′ |
| mTOR | Sense Antisense | 5′-CACATCACTCCCTTCACCA-3′ 5′-GCAACAACGGCTTTCCAC-3′ |
| MuRF-1 | Sense Antisense | 5′-ATCTAGCCTGATTCCTGATGGA-3′ 5′ACCACAGGCTTGGTAAACATCT3′ |
| Foxo-3a | Sense Antisense | 5ACCTTCGTCTCTGAACCTTG–3′ 5′AGTGTGACACGGAAGAGAAGGT3′ |
| Smad 2/3 | Sense Antisense | 5GTCCCAACCCAGGAGATCAT–3′ 5′CCACGATGGACGTAAGGGAG3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurera, M.J.M.S.; Nagahawatta, D.P.; Liyanage, N.M.; Jayawardhana, H.H.A.C.K.; Dissanayake, D.S.; Lee, H.-G.; Kim, Y.-S.; Kang, S.I.; Jeon, Y.-J. Exploring the Potential of Crassostrea nippona Hydrolysates as Dietary Supplements for Mitigating Dexamethasone-Induced Muscle Atrophy in C2C12 Cells. Mar. Drugs 2024, 22, 113. https://doi.org/10.3390/md22030113
Kurera MJMS, Nagahawatta DP, Liyanage NM, Jayawardhana HHACK, Dissanayake DS, Lee H-G, Kim Y-S, Kang SI, Jeon Y-J. Exploring the Potential of Crassostrea nippona Hydrolysates as Dietary Supplements for Mitigating Dexamethasone-Induced Muscle Atrophy in C2C12 Cells. Marine Drugs. 2024; 22(3):113. https://doi.org/10.3390/md22030113
Chicago/Turabian StyleKurera, M. J. M. S., D. P. Nagahawatta, N. M. Liyanage, H. H. A. C. K. Jayawardhana, D. S. Dissanayake, Hyo-Geun Lee, Young-Sang Kim, Sang In Kang, and You-Jin Jeon. 2024. "Exploring the Potential of Crassostrea nippona Hydrolysates as Dietary Supplements for Mitigating Dexamethasone-Induced Muscle Atrophy in C2C12 Cells" Marine Drugs 22, no. 3: 113. https://doi.org/10.3390/md22030113