Metabolomics-Guided Discovery of New Dimeric Xanthones from Co-Cultures of Mangrove Endophytic Fungi Phomopsis asparagi DHS-48 and Phomopsis sp. DHS-11
Abstract
:1. Introduction
2. Results and Discussions
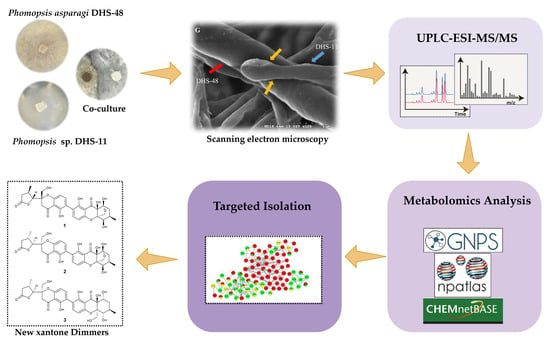
2.1. Morphology of Co-Culture Systems
2.2. Metabolomics Analysis of Large-Scale Fermentation of Co-Cultures
2.3. Structure Elucidation of New Compounds
2.4. Biological Evaluation of Isolated Compounds
3. Materials and Methods
3.1. General Procedures
3.2. Fungal Material
3.3. Preparation of Phomopsis asparagi, Phomopsis sp., Co-Cultivation, and Morphological Observation
3.4. Sample Preparation of Phomopsis asparagi, Phomopsis sp., Co-Culture, and Large-Scale Fermentation and Extracts
3.5. UPLC-ESI-MS/MS Analysis
3.6. Data Processing, Molecular Networking, Dereplication, and Multivariate Data Analysis
3.7. Isolation of Compounds
- Phomoxanthone L (1): yellow amorphous powder (MeOH); [α]20D −20 (c 0.0001, MeOH); UV (MeOH) λmax 205, 251, 361 nm (the absorptions due to aromatic rings); 1H and 13C NMR data, see Table 1; HRESIMS m/z 597.1613 [M − H]− (calcd for C30H29O13 597.1614).
- Phomoxanthone M (2): yellow amorphous powder (MeOH); [α]20D −10 (c 0.0001, MeOH); UV (MeOH) λmax 207, 254, 364 nm (the absorptions due to aromatic rings); 1H and 13C NMR data, see Table 1; HRESIMS m/z 599.1759 [M+H]+ (calcd for C30H31O13 599.1759).
- Phomoxanthone N (3): yellow amorphous powder (MeOH); [α]20D +10 (c 0.0001, MeOH); UV (MeOH) λmax 212, 255, 366 nm (the absorptions due to aromatic rings); 1H and 13C NMR data, see Table 1; HRESIMS m/z 601.1916 [M+H]+ (calcd for C30H33O13 601.1916).
3.8. Theory and Calculation Details
3.9. Cytotoxicity Assay
3.10. Splenocyte Proliferation Assay
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, M.J.; Xu, B.; Guo, Y.W. Unusual Secondary Metabolites from the Mangrove Ecosystems: Structures, Bioactivities, Chemical, and Bio-Syntheses. Mar. Drugs 2022, 20, 535. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Li, G.; Xu, J. Bio-Active Products from Mangrove Ecosystems. Mar. Drugs 2023, 21, 239. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Biomolecules Produced by Mangrove-Associated Microbes. Curr. Med. Chem. 2011, 18, 5224–5266. [Google Scholar] [CrossRef]
- Li, K.; Chen, S.; Pang, X.; Cai, J.; Zhang, X.; Liu, Y.; Zhu, Y.; Zhou, X. Natural Products from Mangrove Sediments-Derived Microbes: Structural Diversity, Bioactivities, Biosynthesis, and Total Synthesis. Eur. J. Med. Chem. 2022, 230, 114117. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.G.; Buchanan, G.O.; Feling, R.H.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. New Cytotoxic Salinosporamides from the Marine Actinomycete Salinispora tropica. J. Org. Chem. 2005, 70, 6196–6203. [Google Scholar] [CrossRef]
- Buckingham, L. EU/3/18/2119. Available online: https://www.ema.europa.eu/en/medicines/human/orphan-designations/eu3182119 (accessed on 8 May 2020).
- Chen, S.; Cai, R.; Liu, Z.; Cui, H.; She, Z. Secondary Metabolites from Mangrove-Associated Fungi: Source, Chemistry and Bioactivities. Nat. Prod. Rep. 2022, 39, 560–595. [Google Scholar] [CrossRef]
- Challis, G.L. Genome Mining for Novel Natural Product Discovery. J. Med. Chem. 2008, 51, 2618–2628. [Google Scholar] [CrossRef]
- Nicault, M.; Zaiter, A.; Dumarcay, S.; Chaimbault, P.; Gelhaye, E.; Leblond, P.; Bontemps, C. Elicitation of Antimicrobial Active Compounds by Streptomyces-Fungus Co-Cultures. Microorganisms 2021, 9, 178. [Google Scholar] [CrossRef]
- Valayil, J.M. Activation of Microbial Silent Gene Clusters: Genomics Driven Drug Discovery Approaches. Biochem. Anal. Biochem. 2016, 5, 276–280. [Google Scholar] [CrossRef]
- Kumar, A.; Sørensen, J.L.; Hansen, F.T.; Arvas, M.; Syed, M.F.; Hassan, L.; Benz, J.P.; Record, E.; Henrissat, B.; Pöggeler, S.; et al. Genome Sequencing and Analyses of Two Marine Fungi from the North Sea Unraveled a Plethora of Novel Biosynthetic Gene Clusters. Sci. Rep. 2018, 8, 10187. [Google Scholar] [CrossRef]
- Gaudêncio, S.P.; Bayram, E.; Lukić Bilela, L.; Cueto, M.; Díaz-Marrero, A.R.; Haznedaroglu, B.Z.; Jimenez, C.; Mandalakis, M.; Pereira, F.; Reyes, F.; et al. Advanced Methods for Natural Products Discovery: Bioactivity Screening, Dereplication, Metabolomics Profiling, Genomic Sequencing, Databases and Informatic Tools, and Structure Elucidation. Mar. Drugs 2023, 21, 308. [Google Scholar] [CrossRef] [PubMed]
- Schroeckh, V.; Scherlach, K.; Nützmann, H.-W.; Shelest, E.; Schmidt-Heck, W.; Schuemann, J.; Martin, K.; Hertweck, C.; Brakhage, A.A. Intimate Bacterial–Fungal Interaction Triggers Biosynthesis of Archetypal Polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 2009, 106, 14558–14563. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite Induction via Microorganism Co-Culture: A Potential Way to Enhance Chemical Diversity for Drug Discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef] [PubMed]
- Knowles, S.L.; Raja, H.A.; Roberts, C.D.; Oberlies, N.H. Fungal–Fungal Co-Culture: A Primer for Generating Chemical Diversity. Nat. Prod. Rep. 2022, 39, 1557–1573. [Google Scholar] [CrossRef] [PubMed]
- Caudal, F.; Tapissier-Bontemps, N.; Edrada-Ebel, R.A. Impact of Co-Culture on the Metabolism of Marine Microorganisms. Mar. Drugs 2022, 20, 153. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Chen, G.; Wu, J.; Pan, J. Structure Revision and Cytotoxic Activity of Marinamide and Its Methyl Ester, Novel Alkaloids Produced by Co-Cultures of Two Marine-Derived Mangrove Endophytic Fungi. Nat. Prod. Res. 2013, 27, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Lu, Y.; Feng, Z.; Luo, S.; Li, C. A New Nonadride Derivative from the Co-Culture Broth of Two Mangrove Fungi. Chem. Nat. Compd. 2017, 53, 691–693. [Google Scholar] [CrossRef]
- Li, C.; Zhang, J.; Zhong, J.; He, B.; Li, R.; Liang, Y.; Lin, Y.; Zhou, S. Isolation and identication of the metabolites from the mixed fermentation broth of two mangrove endophytic fungi. J. South China Agric. Univ. 2011, 32, 117–119. [Google Scholar] [CrossRef]
- Li, C.; Zhang, J.; Shao, C.; Ding, W.; She, Z.; Lin, Y. A new xanthone derivative from the co-culture broth of two marine fungi (strain no. E33 and K38). Chem. Nat. Compd. 2011, 47, 382–384. [Google Scholar] [CrossRef]
- Shao, C.L.; Guo, Z.Y.; Xia, X.K.; Liu, Y.; Huang, Z.-J.; She, Z.-G.; Lin, Y.-C.; Zhou, S.-N. Five Nitro-Phenyl Compounds from the South China Sea Mangrove Fungus. J. Asian Nat. Prod. Res. 2007, 9, 643–648. [Google Scholar] [CrossRef]
- Shao, C.; Hu, G.; Zhang, X.; Wang, C.; She, Z.; Lin, Y. A New Protocatechuic Acid Derivative from the Mangrove Endophytic Fungus B60. Acta Sci. Nat. Univ. Sunyatseni 2008, 5, 133–134. [Google Scholar]
- Huang, H.; She, Z.; Lin, Y.; Vrijmoed, L.L.P.; Lin, W. Cyclic Peptides from an Endophytic Fungus Obtained from a Mangrove Leaf (Kandelia candel). J. Nat. Prod. 2007, 70, 1696–1699. [Google Scholar] [CrossRef]
- Li, C.; Cox, D.; Huang, S.; Ding, W. Two New Cyclopeptides from the Co-Culture Broth of Two Marine Mangrove Fungi and Their Antifungal Activity. Pharmacogn. Mag. 2014, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Covington, B.C.; McLean, J.A.; Bachmann, B.O. Comparative Mass Spectrometry-Based Metabolomics Strategies for the Investigation of Microbial Secondary Metabolites. Nat. Prod. Rep. 2017, 34, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Yuliana, N.D.; Jahangir, M.; Verpoorte, R.; Choi, Y.H. Metabolomics for the Rapid Dereplication of Bioactive Compounds from Natural Sources. Phytochem. Rev. 2013, 12, 293–304. [Google Scholar] [CrossRef]
- Oppong-Danquah, E.; Parrot, D.; Blümel, M.; Labes, A.; Tasdemir, D. Molecular Networking-Based Metabolome and Bioactivity Analyses of Marine-Adapted Fungi Co-Cultivated with Phytopathogens. Front. Microbiol. 2018, 9, 2072. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-Y.; Shen, X.-T.; Yuan, X.-J.; Zhou, Y.-M.; Fan, H.; Zhu, L.-P.; Du, F.-Y.; Sadilek, M.; Yang, J.; Qiao, B.; et al. Metabolomics Investigation of an Association of Induced Features and Corresponding Fungus during the Co-Culture of Trametes Versicolor and Ganoderma applanatum. Front. Microbiol. 2018, 8, 2647. [Google Scholar] [CrossRef]
- Stierle, A.A.; Stierle, D.B.; Decato, D.; Priestley, N.D.; Alverson, J.B.; Hoody, J.; McGrath, K.; Klepacki, D. The Berkeleylactones, Antibiotic Macrolides from Fungal Coculture. J. Nat. Prod. 2017, 80, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Quinn, R.A.; Nothias, L.-F.; Vining, O.; Meehan, M.; Esquenazi, E.; Dorrestein, P.C. Molecular Networking as a Drug Discovery, Drug Metabolism, and Precision Medicine Strategy. Trends Pharmacol. Sci. 2017, 38, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; Van Der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.M.; et al. Mass Spectral Molecular Networking of Living Microbial Colonies. Proc. Natl. Acad. Sci. USA 2012, 109, E1743–E1752. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Um, S.; Seibel, E.; Schalk, F.; Balluff, S.; Beemelmanns, C. Targeted Isolation of Saalfelduracin B–D from Amycolatopsis saalfeldensis Using LC-MS/MS-Based Molecular Networking. J. Nat. Prod. 2021, 84, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Vallet, M.; Vanbellingen, Q.P.; Fu, T.; Le Caer, J.-P.; Della-Negra, S.; Touboul, D.; Duncan, K.R.; Nay, B.; Brunelle, A.; Prado, S. An Integrative Approach to Decipher the Chemical Antagonism between the Competing Endophytes Paraconiothyrium variabile and Bacillus subtilis. J. Nat. Prod. 2017, 80, 2863–2873. [Google Scholar] [CrossRef] [PubMed]
- Chagas, F.O.; Caraballo-Rodríguez, A.M.; Dorrestein, P.C.; Pupo, M.T. Expanding the Chemical Repertoire of the Endophyte Streptomyces albospinus RLe7 Reveals Amphotericin B as an Inducer of a Fungal Phenotype. J. Nat. Prod. 2017, 80, 1302–1309. [Google Scholar] [CrossRef]
- Xu, Z.; Xiong, B.; Xu, J. Chemical Investigation of Secondary Metabolites Produced by Mangrove Endophytic Fungus Phyllosticta capitalensis. Nat. Prod. Res. 2021, 35, 1561–1565. [Google Scholar] [CrossRef]
- Hemberger, Y.; Xu, J.; Wray, V.; Proksch, P.; Wu, J.; Bringmann, G. Pestalotiopens A and B: Stereochemically Challenging Flexible Sesquiterpene-Cyclopaldic Acid Hybrids from Pestalotiopsis sp. Chem. Eur. J. 2013, 19, 15556–15564. [Google Scholar] [CrossRef]
- Deng, Q.; Li, G.; Sun, M.; Yang, X.; Xu, J. A New Antimicrobial Sesquiterpene Isolated from Endophytic Fungus Cytospora sp. from the Chinese Mangrove Plant Ceriops tagal. Nat. Prod. Res. 2020, 34, 1404–1408. [Google Scholar] [CrossRef]
- Sun, M.Y.; Zhou, D.D.; Wu, J.W.; Zhou, J.; Xu, J. Sdy-1 Executes Antitumor Activity in HepG2 and HeLa Cancer Cells by Inhibiting the Wnt/β-Catenin Signaling Pathway. Mar. Drugs 2022, 20, 125. [Google Scholar] [CrossRef]
- Tan, M.; Xu, X.; Zhang, W.; Wu, F.; Bo, X.; Qin, F.; Ju, S.; Song, Z.; Yang, T.; Li, J.; et al. Isolation and Insecticidal Activities of New Cyclic Peptides from Mangrove Endophytic Fungus Aspergillus Sp. GXNU-4QQY1a. Fitoterapia 2023, 171, 105693. [Google Scholar] [CrossRef]
- Xu, J.; Kjer, J.; Sendker, J.; Wray, V.; Guan, H.; Edrada, R.; Lin, W.H.; Wu, J.; Proksch, P. Chromones from the Endophytic Fungus Pestalotiopsis sp. Isolated from the Chinese Mangrove Plant Rhizophora mucronata. J. Nat. Prod. 2009, 72, 662–665. [Google Scholar] [CrossRef]
- Zhou, J.; Li, G.; Deng, Q.; Zheng, D.Y.; Yang, X.B.; Xu, J. Cytotoxic Constituents from the Mangrove Endophytic Pestalotiopsis sp. Induce G0/G1 Cell Cycle Arrest and Apoptosis in Human Cancer Cells. Nat. Prod. Res. 2018, 32, 2968–2972. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.W.; Sun, C.X.; Feng, Z.; Zhang, X.X.; Xu, J. Four New Chromones from the Endophytic Fungus Phomopsis asparagi DHS-48 Isolated from the Chinese Mangrove Plant Rhizophora mangle. Mar. Drugs 2021, 19, 348. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Y.; Wu, X.; Li, G.; Feng, Z.; Xu, J. Pestalotiopisorin B, a New Isocoumarin Derivative from the Mangrove Endophytic Fungus Pestalotiopsis sp. HHL101. Nat. Prod. Res. 2020, 34, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Li, G.; Deng, Q.; Xu, Z.Y.; Cen, J.R.; Xu, J. Vomifoliol Isolated from Mangrove Plant Ceriops tagal Inhibits the NFAT Signaling Pathway with CN as the Target Enzyme In Vitro. Bioorganic Med. Chem. Lett. 2021, 48, 128235. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, X.; Wu, J.; Wei, C.; Feng, T.; Zhou, D.; Wen, Z.; Xu, J. Immunosuppressive cytochalasins from the mangrove endophytic fungus Phomopsis asparagi DHS-48. Mar. Drugs 2022, 20, 526. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Wei, C.; Deng, X.; Chen, D.; Wen, Z.; Xu, J. Epigenetic manipulation induced production of immunosuppressive chromones and cytochalasins from the mangrove endophytic fungus Phomopsis asparagi DHS-48. Mar. Drugs 2022, 20, 616. [Google Scholar] [CrossRef]
- Stockert, J.C.; Blázquez-Castro, A.; Cañete, M.; Horobin, R.W.; Villanueva, Á. MTT Assay for Cell Viability: Intracellular Localization of the Formazan Product Is in Lipid Droplets. Acta Histochem. 2012, 114, 785–796. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, B.; Chen, D.; Deng, X.; Yuan, J.; Zhang, S.; Xiong, Z.; Xu, J. New Isocoumarin and Pyrone Derivatives from the Chinese Mangrove Plant Rhizophora mangle-Associated Fungus Phomopsis sp. DHS-11. Molecules 2023, 28, 3756. [Google Scholar] [CrossRef]
- Bertrand, S.; Schumpp, O.; Bohni, N.; Bujard, A.; Azzollini, A.; Monod, M.; Gindro, K.; Wolfender, J.L. Detection of metabolite induction in fungal co-cultures on solid media by high-throughput differen tial ultra-high pressure liquid chromatography-time-of-flight mass spectrometry of ingerprinting. J. Chromatogr. A 2013, 1292, 219–228. [Google Scholar] [CrossRef]
- Prabhu, G.; Bhat, D.; Bhat, R.M.; Selvaraj, S. A Critical Look at Bioproducts Co-Cultured Under Solid State Fermentation and Their Challenges and Industrial Applications. Waste Biomass Valorization 2022, 13, 3095–3111. [Google Scholar] [CrossRef]
- Cartabia, M.; Girometta, C.E.; Milanese, C.; Baiguera, R.M.; Buratti, S.; Branciforti, D.S.; Vadivel, D.; Girella, A.; Babbini, S.; Savino, E.; et al. Collection and Characterization of Wood Decay Fungal Strains for Developing Pure Mycelium Mats. J. Fungi 2021, 7, 1008. [Google Scholar] [CrossRef] [PubMed]
- Dashti, Y.; Grkovic, T.; Abdelmohsen, U.R.; Hentschel, U.; Quinn, R.J. Actinomycete Metabolome Induction/Suppression with N-Acetylglucosamine. J. Nat. Prod. 2017, 80, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Yuan, J.; Huang, X.; Wen, W.; Zhu, X.; Liu, Y.; Li, H.; Lu, Y.; He, L.; Tan, H.; et al. New Dimeric Members of the Phomoxanthone Family: Phomolactonexanthones A, B and Deacetylphomoxanthone C Isolated from the Fungus Phomopsis sp. Mar. Drugs 2013, 11, 4961–4972. [Google Scholar] [CrossRef] [PubMed]
- Stewart, S.; Ivy, M.A.; Anslyn, E.V. The Use of Principal Component Analysis and Discriminant Analysis in Differential Sensing Routines. Chem. Soc. Rev. 2014, 43, 70–84. [Google Scholar] [CrossRef]
- Hamed, A.A.; Soldatou, S.; Qader, M.M.; Arjunan, S.; Miranda, K.J.; Casolari, F.; Pavesi, C.; Diyaolu, O.A.; Thissera, B.; Eshelli, M.; et al. Screening Fungal Endophytes Derived from Under-Explored Egyptian Marine Habitats for Antimicrobial and Antioxidant Properties in Factionalised Textiles. Microorganisms 2020, 8, 1617. [Google Scholar] [CrossRef]
- Crüsemann, M.; O’Neill, E.C.; Larson, C.B.; Melnik, A.V.; Floros, D.J.; Da Silva, R.R.; Jensen, P.R.; Dorrestein, P.C.; Moore, B.S. Prioritizing Natural Product Diversity in a Collection of 146 Bacterial Strains Based on Growth and Extraction Protocols. J. Nat. Prod. 2017, 80, 588–597. [Google Scholar] [CrossRef]
- Zhou, D.D.; Feng, T.; Xu, J. Mangrove endophytic fungi-derived dicerandrol A and its inhibitory effects and preliminary mechanism on HepG2 cells. Chin. J. Antibiot. 2022, 47, 481–487. [Google Scholar] [CrossRef]
- Combès, A.; Ndoye, I.; Bance, C.; Bruzaud, J.; Djediat, C.; Dupont, J.; Nay, B.; Prado, S. Chemical Communication between the Endophytic Fungus Paraconiothyrium Variabile and the Phytopathogen Fusarium Oxysporum. PLoS ONE 2012, 7, e47313. [Google Scholar] [CrossRef]
- Wang, Y.; Glukhov, E.; He, Y.; Liu, Y.; Zhou, L.; Ma, X.; Hu, X.; Hong, P.; Gerwick, W.H.; Zhang, Y. Secondary Metabolite Variation and Bioactivities of Two Marine Aspergillus Strains in Static Co-Culture Investigated by Molecular Network Analysis and Multiple Database Mining Based on LC-PDA-MS/MS. Antibiotics 2022, 11, 513. [Google Scholar] [CrossRef]
- Oppong-Danquah, E.; Blümel, M.; Scarpato, S.; Mangoni, A.; Tasdemir, D. Induction of Isochromanones by Co-Cultivation of the Marine Fungus Cosmospora sp. and the Phytopathogen Magnaporthe oryzae. Int. J. Mol. Sci. 2022, 23, 782. [Google Scholar] [CrossRef]
- Liu, S.; Wang, T.; Lu, Q.; Li, F.; Wu, G.; Jiang, Z.; Habden, X.; Liu, L.; Zhang, X.; Lukianov, D.A.; et al. Bioprospecting of Soil-Derived Actinobacteria Along the Alar-Hotan Desert Highway in the Taklamakan Desert. Front. Microbiol. 2021, 12, 604999. [Google Scholar] [CrossRef] [PubMed]
- Van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [PubMed]








| Position | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δC Type | δH (J in Hz) | δC Type | δH (J in Hz) | δC Type | δH (J in Hz) | |
| 1 | 159.9, C | 161.5, C | 158.9, C | |||
| 2 | 118.4, C | 119.0, C | 116.1, C | |||
| 3 | 141.8, CH | 7.47, d, 8.5 | 143.5, CH | 7.55, d, 8.4 | 139.6, CH | 7.39, d, 8.5 |
| 4 | 107.4, CH | 6.62, d, 8.5 | 108.4, CH | 6.59, d, 8.4 | 107.9, CH | 6.55, d, 8.5 |
| 4a | 158.2, C | 159.4, C | 158.2, C | |||
| 5 | 76.4, C | 4.11, d, 4.1 | 77.8, C | 4.10, d, 3.8 | 72.3, C | 4.16, s |
| 6 | 29.2, C | 2.33, m | 30.5, C | 2.30, m | 27.4, C | 2.11, m |
| 7 | 37.1, CH2 | Ha 2.09, d, 13.8 | 38.5, CH2 | Ha 2.10, t, 13.5 | 31.2, CH2 | Ha 1.87, t, 10.9 |
| Hb 1.69, dd, 13.8, 5.8 | Hb 1.68, dd, 13.5, 5.8 | Hb 1.41, d, 10.9 | ||||
| 8 | 106.5, C | 107.9, C | 66.4, CH | 4.23, s | ||
| 8a | 71.8, C | 73.0, C | 74.8, C | |||
| 9 | 199.76, C | 199.27, C | 194.9, C | |||
| 9a | 108.6, C | 107.0, C | 106.85, C | |||
| 10a | 81.9, C | 83.2, C | 84.4, C | |||
| 11 | 14.7, CH3 | 1.12, d, 6.8 | 16.1, CH3 | 1.12, d, 6.8 | 17.9, CH3 | 0.98, d, 6.5 |
| 12 | 64.6, CH2 | Ha 3.89, d, 9.7 | 66.0, CH2 | Ha 3.95, d, 9.6 | 58.3, CH2 | Ha 4.12, d, 13.2 |
| Hb 3.83, d, 9.7 | Hb 3.78, d, 9.6 | Hb 3.57, d, 13.2 | ||||
| 1′ | 161.1, C | 159.97, C | 158.0, C | |||
| 2′ | 115.5, C | 117.8, C | 116.3, C | |||
| 3′ | 139.6, CH | 7.36, d, 8.5 | 141.7, CH | 7.45, d, 8.4 | 140.0, CH | 7.40, d, 8.5 |
| 4′ | 108.6, CH | 6.52, d, 8.5 | 108.0, CH | 6.49, d, 8.4 | 106.67, CH | 6.47, d, 8.5 |
| 4a′ | 156.2, C | 160.4, C | 158.8, C | |||
| 5′ | 84.7, CH | 4.30, d, 3.4 | 88.6, CH | 4.43, d, 4.0 | 86.6, CH | 4.36, d, 4.2 |
| 6′ | 28.7, CH | 2.61, m | 31.0, CH | 2.86, m | 29.2, CH | 2.80, m |
| 7′ | 35.6, CH2 | Ha 2.17, dd, 17.7, 9.0 | 37.3, CH2 | Ha 2.88, m | 35.9, CH2 | Ha 2.77, m |
| Hb 1.95, dd, 17.7, 9.2 | Hb 2.28, m | Hb 2.23, m | ||||
| 8′ | 177.3, C | 178.7, C | 176.1, C | |||
| 8a′ | 37.3, CH2 | Ha 3.21, m | 37.8, CH2 | 2.88, m | 37.2, CH2 | Ha, 3.15, d, 17.1 |
| Hb 2.92, m | Hb 2.91, d, 17.1 | |||||
| 9′ | 197.94, C | 198.46, C | 197.5, C | |||
| 9a′ | 107.0, C | 108.5, C | 106.92, C | |||
| 10a′ | 83.6, C | 84.9, C | 84.0, C | |||
| 11′ | 19.6, CH3 | 1.09, d, 7.1 | 20.8, CH3 | 1.23, d, 5.9 | 20.1, CH3 | 1.13, d, 6.3 |
| 12′ | 61.0, CH2 | Ha 3.82, d, 11.6 | 63.1, CH2 | 3.82, d, 2.1 | 62.3, CH2 | 3.67, d, 4.1 |
| Hb 3.71, d, 11.6 | ||||||
| Compound | IC50 (µM) a | |
|---|---|---|
| HepG2 | Hela | |
| 1 | 53.72 ± 1.22 | 69.53 ± 2.25 |
| 2 | 50.25 ± 1.08 | 67.66 ± 1.89 |
| 3 | 67.32 ± 0.88 | 87.32 ± 0.98 |
| 4 | 45.69 ± 0.69 | 41.25 ± 0.26 |
| 5 | 48.75 ± 0.85 | 48.11 ± 1.00 |
| 6 | 48.10 ± 0.26 | 47.22 ± 0.34 |
| 7 | 4.83 ± 0.22 | 18.96 ± 0.88 |
| 8 | 13.99 ± 1.13 | 23.42 ± 2.55 |
| 9 | 12.06 ± 0.55 | 20.36 ± 1.99 |
| Adriamycin b | \ | 0.88 ± 0.71 |
| Fluorouracil c | 179.03 ± 25.82 | \ |
| Compound | IC50 (µM) a | |
|---|---|---|
| ConA-Induced T-Cell Proliferation | LPS-Induced B-Cell Proliferation | |
| 1 | 55.53 ± 0.93 | 89.27 ± 2.25 |
| 2 | 60.25 ± 1.58 | 87.66 ± 2.76 |
| 3 | 75.75 ± 1.78 | 102.65 ± 1.38 |
| 4 | 44.84 ± 1.26 | 77.76 ± 1.47 |
| 5 | 55.71 ± 1.85 | 119.84 ± 1.12 |
| 6 | 57.39 ± 1.24 | 87.72 ± 0.44 |
| 7–9 | - | - |
| cyclosporin A b | 4.39 ± 0.02 | 25.11 ± 0.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Chen, D.; Li, Q.; Feng, T.; Xu, J. Metabolomics-Guided Discovery of New Dimeric Xanthones from Co-Cultures of Mangrove Endophytic Fungi Phomopsis asparagi DHS-48 and Phomopsis sp. DHS-11. Mar. Drugs 2024, 22, 102. https://doi.org/10.3390/md22030102
Wu J, Chen D, Li Q, Feng T, Xu J. Metabolomics-Guided Discovery of New Dimeric Xanthones from Co-Cultures of Mangrove Endophytic Fungi Phomopsis asparagi DHS-48 and Phomopsis sp. DHS-11. Marine Drugs. 2024; 22(3):102. https://doi.org/10.3390/md22030102
Chicago/Turabian StyleWu, Jingwan, Dandan Chen, Qing Li, Ting Feng, and Jing Xu. 2024. "Metabolomics-Guided Discovery of New Dimeric Xanthones from Co-Cultures of Mangrove Endophytic Fungi Phomopsis asparagi DHS-48 and Phomopsis sp. DHS-11" Marine Drugs 22, no. 3: 102. https://doi.org/10.3390/md22030102