Antibacterial Activities and Life Cycle Stages of Asparagopsis armata: Implications of the Metabolome and Microbiome
Abstract
:1. Introduction
2. Results
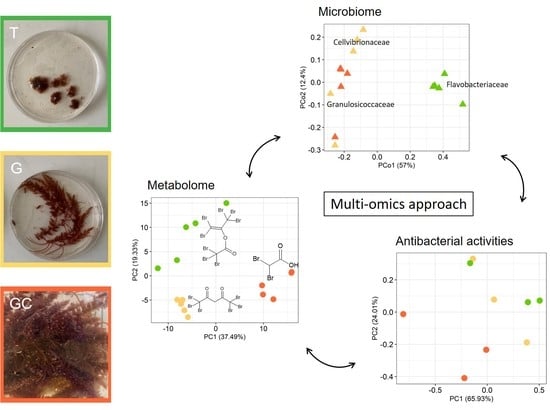
2.1. Variation of the Metabolome of the Three Life Stages
2.2. Variation in the Antibacterial Activity of the Three Life Stages
2.3. Identification of Candidate Molecules Responsible for Antibacterial Activity
2.4. Analysis of Bacterial Communities Associated with A. armata Stages
2.5. Correlation between Metabolite and Bacterial Compositions in Algal Stages
3. Discussion
3.1. The Three Stages of the Life Cycle of A. armata Harboured Distinct Metabolome Compositions
3.2. Highly Halogenated Molecules May Be Responsible for Some Antibacterial Activity
3.3. Microbiota Composition Differs throughout the Three Stages of the Life Cycle of A. armata
3.4. The Overall Analyses Performed Suggest Complex Interactions between the Host and Its Microbiota
4. Materials and Methods
4.1. Chemicals
4.2. Biological Materials
4.3. Algal Extraction
4.4. Metabolomics
4.4.1. Chemical Analyses
4.4.2. Data Treatment
4.4.3. Multivariate and Statistical Analyses
4.5. Antibacterial Activity
4.6. Bioguided Fractionation and Identification of a Candidate Molecule Responsible for the Antibacterial Activity
4.7. Metabarcoding
4.7.1. DNA Extraction, PCR and Sequencing
4.7.2. Sequence Analyses
4.7.3. Multivariate and Statistical Analyses
4.8. Multi-Omics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kladi, M.; Vagias, C.; Roussis, V. Volatile Halogenated Metabolites from Marine Red Algae. Phytochem. Rev. 2004, 3, 337–366. [Google Scholar] [CrossRef]
- Paul, C.; Pohnert, G. Production and Role of Volatile Halogenated Compounds from Marine Algae. Nat. Prod. Rep. 2011, 28, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Egan, S.; Harder, T.; Burke, C.; Steinberg, P.; Kjelleberg, S.; Thomas, T. The Seaweed Holobiont: Understanding Seaweed–Bacteria Interactions. FEMS Microbiol. Rev. 2013, 37, 462–476. [Google Scholar] [CrossRef] [Green Version]
- Dworjanyn, S.A.; De Nys, R.; Steinberg, P.D. Localisation and Surface Quantification of Secondary Metabolites in the Red Alga Delisea Pulchra. Mar. Biol. 1999, 133, 727–736. [Google Scholar] [CrossRef]
- Maximilien, R.; de Nys, R.; Holmström, C.; Gram, L.; Givskov, M.; Crass, K.; Kjelleberg, S.; Steinberg, P.D. Chemical Mediation of Bacterial Surface Colonisation by Secondary Metabolites from the Red Alga Delisea Pulchra. Aquat. Microb. Ecol. 1998, 15, 233–246. [Google Scholar] [CrossRef]
- Paul, N.A.; de Nys, R.; Steinberg, P.D. Chemical Defence against Bacteria in the Red Alga Asparagopsis armata: Linking Structure with Function. Mar. Ecol. Prog. Ser. 2006, 306, 87–101. [Google Scholar] [CrossRef] [Green Version]
- Nylund, G.M.; Cervin, G.; Persson, F.; Hermansson, M.; Steinberg, P.D.; Pavia, H. Seaweed Defence against Bacteria: A Poly-Brominated 2-Heptanone from the Red Alga Bonnemaisonia Hamifera Inhibits Bacterial Colonisation. Mar. Ecol. Prog. Ser. 2008, 369, 39–50. [Google Scholar] [CrossRef]
- Lane, A.L.; Stout, E.P.; Lin, A.-S.; Prudhomme, J.; Le Roch, K.; Fairchild, C.R.; Franzblau, S.G.; Hay, M.E.; Aalbersberg, W.; Kubanek, J. Antimalarial Bromophycolides J-Q from the Fijian Red Alga Callophycus Serratus. J. Org. Chem. 2009, 74, 2736–2742. [Google Scholar] [CrossRef] [Green Version]
- Burreson, B.J.; Moore, R.E.; Roller, P. Haloforms in the Essential Oil of the Alga Asparagopsis taxiformis (Rhodophyta). Tetrahedron Lett. 1975, 16, 473–476. [Google Scholar] [CrossRef]
- Burreson, B.J.; Moore, R.E.; Roller, P.P. Volatile Halogen Compounds in the Alga Asparagopsis taxiformis (Rhodophyta). J. Agric. Food Chem. 1976, 24, 856–861. [Google Scholar] [CrossRef]
- McConnell, O.; Fenical, W. Halogen Chemistry of the Red Alga Asparagopsis. Phytochemistry 1977, 16, 367–374. [Google Scholar] [CrossRef]
- Woolard, F.X.; Moore, R.E.; Roller, P.P. Halogenated Acetamides, but-3-En-2-Ols, and Isopropanols from Asparagopsis taxiformis (Delile) Trev. Tetrahedron 1976, 32, 2843–2846. [Google Scholar] [CrossRef]
- Woolard, F.X.; Moore, R.E.; Roller, P.P. Halogenated Acetic and Acrylic Acids from the Red Alga Asparagopsis taxiformis. Phytochemistry 1979, 18, 617–620. [Google Scholar] [CrossRef]
- Pinto, D.C.; Lesenfants, M.L.; Rosa, G.P.; Barreto, M.C.; Silva, A.; Seca, A.M. GC-and UHPLC-MS Profiles as a Tool to Valorize the Red Alga Asparagopsis armata. Appl. Sci. 2022, 12, 892. [Google Scholar] [CrossRef]
- Thapa, H.R.; Lin, Z.; Yi, D.; Smith, J.E.; Schmidt, E.W.; Agarwal, V. Genetic and Biochemical Reconstitution of Bromoform Biosynthesis in Asparagopsis Lends Insights into Seaweed ROS Enzymology. ACS Chem. Biol. 2020, 15, 1662–1670. [Google Scholar] [CrossRef]
- Bruneau, Y.; Codomier, L.; Combaut, G.; Teste, J. Etude Comparative Des Composés Halogénés Du Falkenbergia Rufolanesa (HARV.) SCHMITZ et de l’Asparagopsis armata (HARV.): Rhodophycees Bonnemaisoniales. Compte Rendus L’académie Sci. À Paris 1978, 286, 603–605. [Google Scholar]
- Vergés, A.; Paul, N.A.; Steinberg, P.D. Sex and Life-History Stage Alter Herbivore Responses to a Chemically Defended Red Alga. Ecology 2008, 89, 1334–1343. [Google Scholar] [CrossRef] [PubMed]
- Greff, S.; Zubia, M.; Genta-Jouve, G.; Massi, L.; Perez, T.; Thomas, O.P. Mahorones, Highly Brominated Cyclopentenones from the Red Alga Asparagopsis taxiformis. J. Nat. Prod. 2014, 77, 1150–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansemir, A.; Blume, M.; Schröder, S.; Lindequist, U. Screening of Cultivated Seaweeds for Antibacterial Activity against Fish Pathogenic Bacteria. Aquaculture 2006, 252, 79–84. [Google Scholar] [CrossRef]
- Salvador, N.; Gómez Garreta, M.A.; Lavelli, L.; Ribera Siguán, M.A. Antimicrobial Activity of Iberian Macroalgae. Sci. Mar. 2007, 71, 101–113. [Google Scholar] [CrossRef] [Green Version]
- Lemay, M.A.; Martone, P.T.; Hind, K.R.; Lindstrom, S.C.; Wegener Parfrey, L. Alternate Life History Phases of a Common Seaweed Have Distinct Microbial Surface Communities. Mol. Ecol. 2018, 27, 3555–3568. [Google Scholar] [CrossRef] [PubMed]
- Aires, T.; Serrão, E.A.; Engelen, A.H. Host and Environmental Specificity in Bacterial Communities Associated to Two Highly Invasive Marine Species (Genus Asparagopsis). Front. Microbiol. 2016, 7, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horta, A.; Alves, C.; Pinteus, S.; Lopes, C.; Fino, N.; Silva, J.; Ribeiro, J.; Rodrigues, D.; Francisco, J.; Rodrigues, A. Identification of Asparagopsis armata-Associated Bacteria and Characterization of Their Bioactive Potential. MicrobiologyOpen 2019, 8, e00824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parchemin, C.; Raviglione, D.; Ghosson, H.; Salvia, M.-V.; Goossens, C.; Sasal, P.; Faliex, E.; Tapissier-Bontemps, N. Development of a Multiblock Metabolomics Approach to Explore Metabolite Variations of Two Algae of the Genus Asparagopsis Linked to Interspecies and Temporal Factors. Algal Res. 2023, 76, 103138. [Google Scholar] [CrossRef]
- Reverter, M.; Tapissier-Bontemps, N.; Banaigs, B.; Sasal, P.; Calvayrac, C.; Mazzitelli, J.-Y.; Tintillier, F. Composé Antibactérien Et Antiparasitaire. French Patent No. FR3117734, 23 June 2022. [Google Scholar]
- Félix, R.; Dias, P.; Félix, C.; Cerqueira, T.; Andrade, P.B.; Valentão, P.; Lemos, M.F. The Biotechnological Potential of Asparagopsis armata: What Is Known of Its Chemical Composition, Bioactivities and Current Market? Algal Res. 2021, 60, 102534. [Google Scholar] [CrossRef]
- Ponte, J.M.; Seca, A.M.; Barreto, M.C. Asparagopsis Genus: What We Really Know About Its Biological Activities and Chemical Composition. Molecules 2022, 27, 1787. [Google Scholar] [CrossRef]
- Zanolla, M.; Carmona, R.; Mata, L.; De la Rosa, J.; Sherwood, A.; Barranco, C.N.; Muñoz, A.R.; Altamirano, M. Concise Review of the Genus Asparagopsis Montagne, 1840. J. Appl. Phycol. 2022, 34, 1–17. [Google Scholar] [CrossRef]
- Sugano, M.; Sato, A.; Nagak, H.; Yoshiok, S.; Shiraki, T.; Horikoshi, H. Aldose Reductase Inhibitors from the Red Alga, Asparagopsis taxiformis. Tetrahedron Lett. 1990, 31, 7015–7016. [Google Scholar] [CrossRef]
- Zhu, P.; Li, D.; Yang, Q.; Su, P.; Wang, H.; Heimann, K.; Zhang, W. Commercial Cultivation, Industrial Application, and Potential Halocarbon Biosynthesis Pathway of Asparagopsis sp. Algal Res. 2021, 56, 102319. [Google Scholar] [CrossRef]
- Butler, A.; Carter-Franklin, J.N. The Role of Vanadium Bromoperoxidase in the Biosynthesis of Halogenated Marine Natural Products. Nat. Prod. Rep. 2004, 21, 180–188. [Google Scholar] [CrossRef]
- Theiler, R.; Cook, J.C.; Hager, L.P.; Siuda, J.F. Halohydrocarbon Synthesis by Bromoperoxidase. Science 1978, 202, 1094–1096. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, R.; Ozgur, R.; Higashi, Y.; Takechi, K.; Takano, H.; Takio, S. Preferential Expression of a Bromoperoxidase in Sporophytes of a Red Alga, Pyropia Yezoensis. Mar. Biotechnol. 2015, 17, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.A.; Hamilton, J.T.; Dring, M.J.; Harper, D.B. Do Vesicle Cells of the Red Alga Asparagopsis (Falkenbergia Stage) Play a Role in Bromocarbon Production? Chemosphere 2003, 52, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Campbell, A.; Patwary, Z.P.; Wang, T.; Webb, J.; Zuccarello, G.C.; Wegner, A.; Heyne, D.; McKinnie, L.; Pascelli, C. Asparagopsis taxiformis Lineage 6 Genome; University of the Sunshine Coast: Sunshine Coast, Australia, 2022. [Google Scholar]
- Wemheuer, F.; Taylor, J.A.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of Habitat-Specific Functional Profiles and Functional Redundancy Based on 16S RRNA Gene Sequences. Environ. Microbiome 2020, 15, 11. [Google Scholar] [CrossRef] [PubMed]
- Tabrett, A.; Horton, M.W. The Influence of Host Genetics on the Microbiome. F1000Research 2020, 9, F1000 Faculty Rev-84. [Google Scholar] [CrossRef]
- Paix, B.; Vieira, C.; Potin, P.; Leblanc, C.; De Clerck, O.; Briand, J.-F.; Culioli, G. French Mediterranean and Atlantic Populations of the Brown Algal Genus Taonia (Dictyotales) Display Differences in Phylogeny, Surface Metabolomes and Epibacterial Communities. Algal Res. 2021, 59, 102452. [Google Scholar] [CrossRef]
- Waaland, J.R. Differences in Carrageenan in Gametophytes and Tetrasporophytes of Red Algae. Phytochemistry 1975, 14, 1359–1362. [Google Scholar] [CrossRef]
- Haslin, C.; Lahaye, M.; Pellegrini, M. Chemical Composition and Structure of Sulphated Water-Soluble Cell-Wall Polysaccharides from the Gametic, Carposporic and Tetrasporic Stages of Asparagopsis armata Harvey (Rhodophyta, Bonnemaisoniaceae). Bot. Mar. 2000, 43, 475–482. [Google Scholar] [CrossRef]
- Mann, A.J.; Hahnke, R.L.; Huang, S.; Werner, J.; Xing, P.; Barbeyron, T.; Huettel, B.; Stüber, K.; Reinhardt, R.; Harder, J. The Genome of the Alga-Associated Marine Flavobacterium Formosa Agariphila KMM 3901T Reveals a Broad Potential for Degradation of Algal Polysaccharides. Appl. Environ. Microbiol. 2013, 79, 6813–6822. [Google Scholar] [CrossRef] [Green Version]
- Michel, G.; Nyval-Collen, P.; Barbeyron, T.; Czjzek, M.; Helbert, W. Bioconversion of Red Seaweed Galactans: A Focus on Bacterial Agarases and Carrageenases. Appl. Microbiol. Biotechnol. 2006, 71, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.; Bordron, P.; Eveillard, D.; Michel, G. Gene Expression Analysis of Zobellia galactanivorans during the Degradation of Algal Polysaccharides Reveals Both Substrate-Specific and Shared Transcriptome-Wide Responses. Front. Microbiol. 2017, 8, 1808. [Google Scholar] [CrossRef] [PubMed]
- Lemay, M.A.; Chen, M.Y.; Mazel, F.; Hind, K.R.; Starko, S.; Keeling, P.J.; Martone, P.T.; Parfrey, L.W. Morphological Complexity Affects the Diversity of Marine Microbiomes. ISME J. 2021, 15, 1372–1386. [Google Scholar] [CrossRef]
- Zanolla, M.; Carmona, R.; De la Rosa, J.; Salvador, N.; Sherwood, A.R.; Andreakis, N.; Altamirano, M. Morphological Differentiation of Cryptic Lineages within the Invasive Genus Asparagopsis (Bonnemaisoniales, Rhodophyta). Phycologia 2014, 53, 233–242. [Google Scholar] [CrossRef]
- Rusconi, R.; Guasto, J.S.; Stocker, R. Bacterial Transport Suppressed by Fluid Shear. Nat. Phys. 2014, 10, 212–217. [Google Scholar] [CrossRef] [Green Version]
- Rusconi, R.; Stocker, R. Microbes in Flow. Curr. Opin. Microbiol. 2015, 25, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Paix, B.; Carriot, N.; Barry-Martinet, R.; Greff, S.; Misson, B.; Briand, J.-F.; Culioli, G. A Multi-Omics Analysis Suggests Links between the Differentiated Surface Metabolome and Epiphytic Microbiota along the Thallus of a Mediterranean Seaweed Holobiont. Front. Microbiol. 2020, 11, 494. [Google Scholar] [CrossRef] [PubMed]
- Grigorian, E.; Groisillier, A.; Thomas, F.; Leblanc, C.; Delage, L. Functional Characterization of a L-2-Haloacid Dehalogenase from Zobellia galactanivorans DsijT Suggests a Role in Haloacetic Acid Catabolism and a Wide Distribution in Marine Environments. Front. Microbiol. 2021, 12, 725997. [Google Scholar] [CrossRef] [PubMed]
- Novak, H.R.; Sayer, C.; Isupov, M.N.; Gotz, D.; Spragg, A.M.; Littlechild, J.A. Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae. FEBS Lett. 2014, 588, 1616–1622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novak, H.R.; Sayer, C.; Panning, J.; Littlechild, J.A. Characterisation of an L-Haloacid Dehalogenase from the Marine Psychrophile Psychromonas Ingrahamii with Potential Industrial Application. Mar. Biotechnol. 2013, 15, 695–705. [Google Scholar] [CrossRef]
- Hudson, J.; Egan, S. Opportunistic Diseases in Marine Eukaryotes: Could Bacteroidota Be the next Threat to Ocean Life? Environ. Microbiol. 2022, 24, 4505–4518. [Google Scholar] [CrossRef]
- Kumar, V.; Zozaya-Valdes, E.; Kjelleberg, S.; Thomas, T.; Egan, S. Multiple Opportunistic Pathogens Can Cause a Bleaching Disease in the Red Seaweed Delisea Pulchra. Environ. Microbiol. 2016, 18, 3962–3975. [Google Scholar] [CrossRef]
- Zozaya-Valdes, E.; Egan, S.; Thomas, T. A Comprehensive Analysis of the Microbial Communities of Healthy and Diseased Marine Macroalgae and the Detection of Known and Potential Bacterial Pathogens. Front. Microbiol. 2015, 6, 146. [Google Scholar] [CrossRef] [Green Version]
- Burke, C.; Steinberg, P.; Rusch, D.; Kjelleberg, S.; Thomas, T. Bacterial Community Assembly Based on Functional Genes Rather than Species. Proc. Natl. Acad. Sci. USA 2011, 108, 14288–14293. [Google Scholar] [CrossRef] [Green Version]
- Giacomoni, F.; Le Corguille, G.; Monsoor, M.; Landi, M.; Pericard, P.; Petera, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.-F.; Jacob, D. Workflow4Metabolomics: A Collaborative Research Infrastructure for Computational Metabolomics. Bioinformatics 2015, 31, 1493–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guitton, Y.; Tremblay-Franco, M.; Le Corguillé, G.; Martin, J.-F.; Pétéra, M.; Roger-Mele, P.; Delabrière, A.; Goulitquer, S.; Monsoor, M.; Duperier, C. Create, Run, Share, Publish, and Reference Your LC–MS, FIA–MS, GC–MS, and NMR Data Analysis Workflows with the Workflow4Metabolomics 3.0 Galaxy Online Infrastructure for Metabolomics. Int. J. Biochem. Cell Biol. 2017, 93, 89–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Der Kloet, F.M.; Bobeldijk, I.; Verheij, E.R.; Jellema, R.H. Analytical Error Reduction Using Single Point Calibration for Accurate and Precise Metabolomic Phenotyping. J. Proteome Res. 2009, 8, 5132–5141. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [Green Version]
- Dührkop, K.; Nothias, L.-F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C. Systematic Classification of Unknown Metabolites Using High-Resolution Fragmentation Mass Spectra. Nat. Biotechnol. 2021, 39, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E. ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.W.; Wang, M.; Leber, C.A.; Nothias, L.-F.; Reher, R.; Kang, K.B.; Van Der Hooft, J.J.; Dorrestein, P.C.; Gerwick, W.H.; Cottrell, G.W. NPClassifier: A Deep Neural Network-Based Structural Classification Tool for Natural Products. J. Nat. Prod. 2021, 84, 2795–2807. [Google Scholar] [CrossRef]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching Molecular Structure Databases with Tandem Mass Spectra Using CSI: FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.A.; Nothias, L.-F.; Ludwig, M.; Fleischauer, M.; Gentry, E.C.; Witting, M.; Dorrestein, P.C.; Dührkop, K.; Böcker, S. Assigning Confidence to Structural Annotations from Mass Spectra with COSMIC. bioRxiv 2021. [Google Scholar] [CrossRef]
- Böcker, S.; Dührkop, K. Fragmentation Trees Reloaded. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [Green Version]
- Böcker, S.; Letzel, M.C.; Lipták, Z.; Pervukhin, A. SIRIUS: Decomposing Isotope Patterns for Metabolite Identification. Bioinformatics 2009, 25, 218–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. MixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castanho, S.; Califano, G.; Soares, F.; Costa, R.; Mata, L.; Pousão-Ferreira, P.; Ribeiro, L. The Effect of Live Feeds Bathed with the Red Seaweed Asparagopsis armata on the Survival, Growth and Physiology Status of Sparus aurata Larvae. Fish Physiol. Biochem. 2017, 43, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Manilal, A.; Selvin, J.; George, S. In Vivo Therapeutic Potentiality of Red Seaweed, Asparagopsis (Bonnemaisoniales, Rhodophyta) in the Treatment of Vibriosis in Penaeus monodon Fabricius. Saudi J. Biol. Sci. 2012, 19, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Reverter, M.; Saulnier, D.; David, R.; Bardon-Albaret, A.; Belliard, C.; Tapissier-Bontemps, N.; Lecchini, D.; Sasal, P. Effects of Local Polynesian Plants and Algae on Growth and Expression of Two Immune-Related Genes in Orbicular Batfish (Platax orbicularis). Fish Shellfish Immunol. 2016, 58, 82–88. [Google Scholar] [CrossRef] [Green Version]
- Thépot, V.; Campbell, A.H.; Rimmer, M.A.; Jelocnik, M.; Johnston, C.; Evans, B.; Paul, N.A. Dietary Inclusion of the Red Seaweed Asparagopsis taxiformis Boosts Production, Stimulates Immune Response and Modulates Gut Microbiota in Atlantic Salmon, Salmo Salar. Aquaculture 2022, 546, 737286. [Google Scholar] [CrossRef]
- Thépot, V.; Campbell, A.H.; Paul, N.; Rimmer, M. Seaweed Dietary Supplements Enhance the Innate Immune Response of the Mottled Rabbitfish, Siganus fuscescens. Fish Shellfish Immunol. 2021, 113, 176–184. [Google Scholar] [CrossRef]
- Thépot, V.; Campbell, A.H.; Rimmer, M.A.; Paul, N.A. Effects of a Seaweed Feed Inclusion on Different Life Stages of the Mottled Rabbitfish Siganus fuscescens. Aquac. Res. 2021, 52, 6626–6640. [Google Scholar] [CrossRef]
- Bauer, A.W. Antibiotic Susceptibility Testing by a Standardized Single Disc Method. Am. J. Clin. Pathol. 1966, 45, 149–158. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Cáceres, M.D.; Legendre, P. Associations between Species and Groups of Sites: Indices and Statistical Inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef]
- Singh, A.; Shannon, C.P.; Gautier, B.; Rohart, F.; Vacher, M.; Tebbutt, S.J.; Lê Cao, K.-A. DIABLO: An Integrative Approach for Identifying Key Molecular Drivers from Multi-Omics Assays. Bioinformatics 2019, 35, 3055–3062. [Google Scholar] [CrossRef]






| GC | G | T | p | |
|---|---|---|---|---|
| Ea | 1.7 ± 0.2 a | 2 ± 0.06 a,b | 2.2 ± 0.1 b | 0.034 |
| Lg * | 0.6 ± 0 | 0.6 ± 0.06 | 0.8 ± 0 | 0.030 |
| Tm | 1.6 ± 0.1 a | 2 ± 0.06 b | 1.9 ± 0.1 a,b | 0.050 |
| Va | 2.0 ± 0.06 | 2.1 ± 0.06 | 2.0 ± 0.06 | 0.141 |
| Vh | 1.4 ± 0.4 | 1.2 ± 0.4 | 1.1 ± 0.4 | 0.610 |
| Yr | 1.4 ± 0.1 a | 1.6 ± 0.1 a,b | 1.7 ± 0.06 b | 0.042 |
| GC | G | T | p | |
|---|---|---|---|---|
| Evenness | 0.78 ± 0.03 | 0.80 ± 0.04 | 0.84 ± 0.02 | 0.069 |
| Chao1 | 513.4 ± 92.4 a | 632.5 ± 235.4 a,b | 920.7 ± 186.6 b | 0.031 |
| Shannon | 4.87 ± 0.3 a | 5.1 ± 0.5 a,b | 5.7 ± 0.3 b | 0.021 |
| Algal Stages | Metabolome | Antibacterial Activity | Microbiota Diversity and Composition | Conclusions and Hypotheses |
|---|---|---|---|---|
| General observation | Significant differences between GC-G, GC-T and G-T | Significant differences between GC-T | Significant Differences between GC-T and G-T | Highly halogenated molecules may be responsible for a part of the antibacterial activity No major covariation between metabolome and microbiota (Mantel) Correlations between specific metabolites and ASVs |
GC | Abundance in C2H2Br2O2, C2H2BrClO2 and C5H4Br2O4 | Lower than T | Lower diversity than T Bacterial community dominated by the same ASVs as G | Distinct metabolome that may be related to the development and protection of reproductive elements |
G | Composition similar to T with abundance in highly brominated molecules (C5H2Br6O2 and C5H2Br5ClO2) | Not significantly different from the two others (but closer to T) | Intermediate diversity between GC and T Bacterial community dominated by the same ASVs as GC | C5H2Br6O2 and C5H2Br5ClO2 may be responsible for a part of the antibacterial activities |
T | Abundance in C5Br8O2 barely detected in GC and G | Higher than GC but not significantly different from G | Diversity higher than GC and G Harbours some specific ASVs not shared with GC or G | Are morphology and annual persistence major factors influencing microbiota composition? Are differences in chemical composition involved in the bacterial diversity or vice versa? Are the greater differences in gene expression involved in specific biosynthetic pathways? |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parchemin, C.; Raviglione, D.; Mejait, A.; Sasal, P.; Faliex, E.; Clerissi, C.; Tapissier-Bontemps, N. Antibacterial Activities and Life Cycle Stages of Asparagopsis armata: Implications of the Metabolome and Microbiome. Mar. Drugs 2023, 21, 363. https://doi.org/10.3390/md21060363
Parchemin C, Raviglione D, Mejait A, Sasal P, Faliex E, Clerissi C, Tapissier-Bontemps N. Antibacterial Activities and Life Cycle Stages of Asparagopsis armata: Implications of the Metabolome and Microbiome. Marine Drugs. 2023; 21(6):363. https://doi.org/10.3390/md21060363
Chicago/Turabian StyleParchemin, Christelle, Delphine Raviglione, Anouar Mejait, Pierre Sasal, Elisabeth Faliex, Camille Clerissi, and Nathalie Tapissier-Bontemps. 2023. "Antibacterial Activities and Life Cycle Stages of Asparagopsis armata: Implications of the Metabolome and Microbiome" Marine Drugs 21, no. 6: 363. https://doi.org/10.3390/md21060363