Ferroptosis Inhibitory Compounds from the Deep-Sea-Derived Fungus Penicillium sp. MCCC 3A00126
Abstract
:1. Introduction
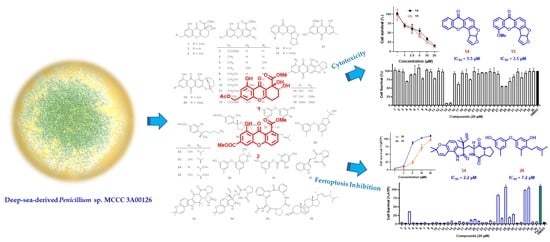
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Biological Material
3.3. Fermentation, Isolation, and Purification
3.4. ECD Calculation
3.5. Cytotoxic Experiment
3.6. Ferroptosis Inhibitory Assay
3.7. DPPH Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2023, 40, 275–325. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2022, 39, 1122–1171. [Google Scholar] [CrossRef] [PubMed]
- Zain Ul Arifeen, M.; Ma, Y.N.; Xue, Y.R.; Liu, C.H. Deep-sea fungi could be the new arsenal for bioactive molecules. Mar. Drugs 2019, 18, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.; Mudassir, S.; Zhang, Z.; Feng, Y.; Chang, Y.; Che, Q.; Gu, Q.; Zhu, T.; Zhang, G.; Li, D. Secondary metabolites from deep-sea derived microorganisms. Curr. Med. Chem. 2020, 27, 6244–6273. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D. Deep-sea natural products. Nat. Prod. Rep. 2008, 25, 1131–1166. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Li, X.; Yang, S.; Li, X.; Wang, B.; Meng, L. Vercytochalasins A and B: Two unprecedented biosynthetically related cytochalasins from the deep-sea-sourced endozoic fungus Curvularia verruculosa. Chin. Chem. Lett. 2023, 34, 107516. [Google Scholar] [CrossRef]
- Yan, L.H.; Li, P.H.; Li, X.M.; Yang, S.Q.; Liu, K.C.; Wang, B.G.; Li, X. Chevalinulins A and B, proangiogenic alkaloids with a spiro[bicyclo[2.2.2]octane-diketopiperazine] skeleton from deep-sea cold-seep-derived fungus Aspergillus chevalieri CS-122. Org. Lett. 2022, 24, 2684–2688. [Google Scholar] [CrossRef]
- Liu, F.A.; Lin, X.; Zhou, X.; Chen, M.; Huang, X.; Yang, B.; Tao, H. Xanthones and quinolones derivatives produced by the deep-sea-derived fungus Penicillium sp. SCSIO Ind16F01. Molecules 2017, 22, 1999. [Google Scholar] [CrossRef] [Green Version]
- Isaka, M.; Palasarn, S.; Kocharin, K.; Saenboonrueng, J. A cytotoxic xanthone dimer from the entomopathogenic fungus Aschersonia sp. BCC 8401. J. Nat. Prod. 2005, 68, 945–946. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Li, H.; Hong, J.; Cho, H.Y.; Bae, K.S.; Kim, M.A.; Kim, D.K.; Jung, J.H. Bioactive metabolites from the sponge-derived fungus Aspergillus versicolor. Arch. Pharm. Res. 2010, 33, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Fredimoses, M.; Zhou, X.; Ai, W.; Tian, X.; Yang, B.; Lin, X.; Liu, J.; Liu, Y. Emerixanthone E, a new xanthone derivative from deep sea fungus Emericella sp. SCSIO 05240. Nat. Prod. Res. 2019, 33, 2088–2094. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.T.; Shan, W.G.; Ying, Y.M.; Ma, L.F.; Liu, W.H.; Zhan, Z.J. Xanthones with α-glucosidase inhibitory activities from Aspergillus versicolor, a fungal endophyte of Huperzia serrata. Helv. Chim. Acta 2015, 98, 148–152. [Google Scholar] [CrossRef]
- Lesch, B.; Brase, S. A short, atom-economical entry to tetrahydroxanthenones. Angew. Chem. Int. Ed. Engl. 2004, 43, 115–118. [Google Scholar] [CrossRef]
- Masters, K.S.; Brase, S. Xanthones from fungi, lichens, and bacteria: The natural products and their synthesis. Chem. Rev. 2012, 112, 3717–3776. [Google Scholar] [CrossRef]
- Verissimo, A.C.S.; Pinto, D.; Silva, A.M.S. Marine-derived xanthone from 2010 to 2021: Isolation, bioactivities and total synthesis. Mar. Drugs 2022, 20, 347. [Google Scholar] [CrossRef]
- Niu, S.; Xie, C.L.; Xia, J.M.; Liu, Q.M.; Peng, G.; Liu, G.M.; Yang, X.W. Botryotins A–H, tetracyclic diterpenoids representing three carbon skeletons from a deep-sea-derived Botryotinia fuckeliana. Org. Lett. 2020, 22, 580–583. [Google Scholar] [CrossRef]
- Xie, C.L.; Liu, Q.; He, Z.H.; Gai, Y.B.; Zou, Z.B.; Shao, Z.Z.; Liu, G.M.; Chen, H.F.; Yang, X.W. Discovery of andrastones from the deep-sea-derived Penicillium allii-sativi MCCC 3A00580 by OSMAC strategy. Bioorg. Chem. 2021, 108, 104671. [Google Scholar] [CrossRef]
- Xie, C.L.; Zhang, D.; Guo, K.Q.; Yan, Q.X.; Zou, Z.B.; He, Z.H.; Wu, Z.; Zhang, X.K.; Chen, H.F.; Yang, X.W. Meroterpenthiazole A, a unique meroterpenoid from the deep-sea-derived Penicillium allii-sativi, significantly inhibited retinoid X receptor (RXR)-α transcriptional effect. Chin. Chem. Lett. 2022, 33, 2057–2059. [Google Scholar] [CrossRef]
- He, Z.H.; Xie, C.L.; Wu, T.; Yue, Y.T.; Wang, C.F.; Xu, L.; Xie, M.M.; Zhang, Y.; Hao, Y.J.; Xu, R.; et al. Tetracyclic steroids bearing a bicyclo[4.4.1] ring system as potent antiosteoporosis agents from the deep-sea-derived fungus Rhizopus sp. W23. J. Nat. Prod. 2023, 86, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Trisuwan, K.; Rukachaisirikul, V.; Kaewpet, M.; Phongpaichit, S.; Hutadilok-Towatana, N.; Preedanon, S.; Sakayaroj, J. Sesquiterpene and xanthone derivatives from the sea fan-derived fungus Aspergillus sydowii PSU-F154. J. Nat. Prod. 2011, 74, 1663–1667. [Google Scholar] [CrossRef] [PubMed]
- Holker, J.S.E.; O’Brien, E.; Simpson, T.J. The structures of some metabolites of Penicillium diversum: α- and β-diversonolic esters. J. Chem. Soc. Perkin Trans. 1 1983, 1365–1368. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Li, A. Total syntheses and structural revision of α- and β-diversonolic esters and total syntheses of diversonol and blennolide C. Angew. Chem. Int. Ed. Engl. 2008, 47, 6579–6582. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Wang, C.; Wei, M.; Gu, Y.; Xia, X.; She, Z.; Lin, Y. Structure elucidation of two new xanthone derivatives from the marine fungus Penicillium sp. (ZZF 32#) from the south china sea. Magn. Reson. Chem. 2008, 46, 1066–1069. [Google Scholar] [PubMed]
- Sun, R.R.; Miao, F.P.; Zhang, J.; Wang, G.; Yin, X.L.; Ji, N.Y. Three new xanthone derivatives from an algicolous isolate of Aspergillus wentii. Magn. Reson. Chem. 2013, 51, 65–68. [Google Scholar] [CrossRef]
- Cui, X.; Zhu, G.; Liu, H.; Jiang, G.; Wang, Y.; Zhu, W. Diversity and function of the antarctic krill microorganisms from Euphausia superba. Sci. Rep. 2016, 6, 36496. [Google Scholar] [CrossRef] [Green Version]
- Hamasaki, T.; Sato, Y.; Hatsuda, Y. Structure of sydowinin A, sydowinin B, and sydowinol, metabolites from Aspergillus sydowi. Agric. Biol. Chem. 2014, 39, 2341–2345. [Google Scholar] [CrossRef]
- Song, X.Q.; Zhang, X.; Han, Q.J.; Li, X.B.; Li, G.; Li, R.J.; Jiao, Y.; Zhou, J.C.; Lou, H.X. Xanthone derivatives from Aspergillus sydowii, an endophytic fungus from the liverwort Scapania ciliata S. Lac and their immunosuppressive activities. Phytochem. Lett. 2013, 6, 318–321. [Google Scholar] [CrossRef]
- Zhu, F.; Lin, Y.C. Three xanthones from a marine-derived mangrove endophytic fungus. Chem. Nat. Compd. 2007, 43, 132–135. [Google Scholar] [CrossRef]
- Shao, C.; She, Z.; Guo, Z.; Peng, H.; Cai, X.; Zhou, S.; Gu, Y.; Lin, Y. 1H and 13C NMR assignments for two anthraquinones and two xanthones from the mangrove fungus (ZSUH-36). Magn. Reson. Chem. 2007, 45, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Graybill, T.L.; Casillas, E.G.; Pal, K.; Townsend, C.A. Silyl triflate-mediated ring-closure and rearrangement in the synthesis of potential bisfuran-containing intermediates of aflatoxin biosynthesis. J. Am. Chem. Soc. 1999, 121, 7729–7746. [Google Scholar] [CrossRef]
- Dou, Y.; Wang, X.; Jiang, D.; Wang, H.; Jiao, Y.; Lou, H.; Wang, X. Metabolites from Aspergillus versicolor, an endolichenic fungus from the lichen Lobaria retigera. Drug Discov. Ther. 2014, 8, 84–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.B.; Zheng, N.; Liang, L.Q.; Zhao, D.M.; Qin, Y.Y.; Li, J.; Yang, R.Y. Secondary metabolites from the endophytic fungus Fusarium equiseti and their antibacterial activities. Chem. Nat. Compd. 2019, 55, 1141–1144. [Google Scholar] [CrossRef]
- Yamazaki, M.; Satoh, Y.; Maebayashi, Y.; Horie, Y. Monoamine oxidase inhibitors from a fungus, Emericella navahoensis. Chem. Pharm. Bull. 1988, 36, 670–675. [Google Scholar] [CrossRef] [Green Version]
- Steyn, P.S.; Vleggaar, R.; Wessels, P.L.; Cole, R.J.; Scott, D.B. Structure and carbon-13 nuclear magnetic resonance assignments of versiconal acetate, versiconol acetate, and versiconol, metabolites from cultures of Aspergillus parasiticus treated with dichlorvos. J. Chem. Soc. Perkin Trans. 1 1979, 451–459. [Google Scholar] [CrossRef]
- Liu, D.; Yan, L.; Ma, L.; Huang, Y.; Pan, X.; Liu, W.; Lv, Z. Diphenyl derivatives from coastal saline soil fungus Aspergillus iizukae. Arch. Pharm. Res. 2015, 38, 1038–1043. [Google Scholar] [CrossRef]
- Lu, Z.; Zhu, H.; Fu, P.; Wang, Y.; Zhang, Z.; Lin, H.; Liu, P.; Zhuang, Y.; Hong, K.; Zhu, W. Cytotoxic polyphenols from the marine-derived fungus Penicillium expansum. J. Nat. Prod. 2010, 73, 911–914. [Google Scholar] [CrossRef]
- Li, X.B.; Zhou, Y.H.; Zhu, R.X.; Chang, W.Q.; Yuan, H.Q.; Gao, W.; Zhang, L.L.; Zhao, Z.T.; Lou, H.X. Identification and biological evaluation of secondary metabolites from the endolichenic fungus Aspergillus versicolor. Chem. Biodivers. 2015, 12, 575–592. [Google Scholar] [CrossRef]
- Sanchez, J.F.; Chiang, Y.M.; Szewczyk, E.; Davidson, A.D.; Ahuja, M.; Elizabeth Oakley, C.; Woo Bok, J.; Keller, N.; Oakley, B.R.; Wang, C.C. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol. Biosyst. 2010, 6, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.Y.; Li, L.; Yang, C.G.; Luo, D.Q.; Zheng, Z.H.; Lu, X.H.; Shi, B.Z. A novel oxybis cresol verticilatin with highly varying degrees of biological activities from the insect pathogenic fungus Paecilomyces verticillatus. J. Asian Nat. Prod. Res. 2014, 16, 1153–1157. [Google Scholar] [CrossRef]
- Hu, S.S.; Jiang, N.; Wang, X.L.; Chen, C.J.; Fan, J.Y.; Wurin, G.G.; Ge, H.M.; Tan, R.X.; Jiao, R.H. Prenylated diphenyl ethers from the mantis-associated fungus Aspergillus versicolor GH-2. Tetrahedron Lett. 2015, 56, 3894–3897. [Google Scholar] [CrossRef]
- Wang, X.; Mou, Y.; Hu, J.; Wang, N.; Zhao, L.; Liu, L.; Wang, S.; Meng, D. Cytotoxic polyphenols from a sponge-associated fungus Aspergillus versicolor Hmp-48. Chem. Biodivers. 2014, 11, 133–139. [Google Scholar] [CrossRef]
- Eamvijarn, A.; Kijjoa, A.; Bruyere, C.; Mathieu, V.; Manoch, L.; Lefranc, F.; Silva, A.; Kiss, R.; Herz, W. Secondary metabolites from a culture of the fungus Neosartorya pseudofischeri and their in vitro cytostatic activity in human cancer cells. Planta Med. 2012, 78, 1767–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grundmann, A.; Li, S.M. Overproduction, purification and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology 2005, 151, 2199–2207. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Kato, H.; Samizo, M.; Nojiri, Y.; Onuki, H.; Hirota, H.; Ohta, T. Notoamides F-K, prenylated indole alkaloids isolated from a marine-derived Aspergillus sp. J. Nat. Prod. 2008, 71, 2064–2407. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Yoshida, T.; Tokue, T.; Nojiri, Y.; Hirota, H.; Ohta, T.; Williams, R.M.; Tsukamoto, S. Notoamides A–D: Prenylated indole alkaloids isolated from a marine-derived fungus, Aspergillus sp. Angew. Chem. Int. Ed. Engl. 2007, 46, 2254–2256. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Gao, H.; Zhang, X.; Wang, S.; Wu, C.; Gu, Q.; Guo, P.; Zhu, T.; Li, D. Psychrophilins E-H and versicotide C, cyclic peptides from the marine-derived fungus Aspergillus versicolor ZLN-60. J. Nat. Prod. 2014, 77, 2218–2223. [Google Scholar] [CrossRef]
- Kobori, M.; Yoshida, M.; Ohnishi-Kameyama, M.; Takei, T.; Shinmoto, H. 5α,8α-Epidioxy-22E-ergosta-6,9(11),22-trien-3β-ol from an edible mushroom suppresses growth of HL60 leukemia and HT29 colon adenocarcinoma cells. Biol. Pharm. Bull. 2006, 29, 755–759. [Google Scholar] [CrossRef] [Green Version]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Monian, P.; Pan, Q.; Zhang, W.; Xiang, J.; Jiang, X. Ferroptosis is an autophagic cell death process. Cell Res. 2016, 26, 1021–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmans, S.; Vanden Berghe, T.; Devisscher, L.; Hassannia, B.; Lyssens, S.; Joossens, J.; Van Der Veken, P.; Vandenabeele, P.; Augustyns, K. Novel ferroptosis inhibitors with improved potency and ADME properties. J. Med. Chem. 2016, 59, 2041–2053. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R. Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell 2022, 185, 2401–2421. [Google Scholar] [CrossRef]
- Feng, H.; Li, F.; Tang, P. Circ_0000745 regulates NOTCH1-mediated cell proliferation and apoptosis in pediatric T-cell acute lymphoblastic leukemia through adsorbing miR-193b-3p. Hematology 2021, 26, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Tan, Q.; Wu, L.; Wu, D.; Xu, J.; Zhou, H.; Gu, Q. Ferroptosis Inhibitory aromatic abietane diterpenoids from Ajuga decumbens and structural revision of two 3,4-epoxy group-containing abietanes. J. Nat. Prod. 2022, 85, 1808–1815. [Google Scholar] [CrossRef] [PubMed]








| No. | 1 | 2 | ||
|---|---|---|---|---|
| δC | δH | δC | δH | |
| 1 | 160.7 C | 161.7 C | ||
| 2 | 109.9 CH | 6.75 s | 111.6 CH | 7.44 s |
| 3 | 145.0 C | 137.6 C | ||
| 4 | 105.5 CH | 6.86 s | 108.1 CH | 7.60 s |
| 4a | 156.1 C | 155.4 C | ||
| 5 | 26.1 CH2 | 2.87 m | 119.6 CH | 7.59 (d, 8.0) |
| 6 | 24.2 CH2 | 2.25 m | 135.5 CH | 7.81 (dd, 8.0, 7.2) |
| 7 | 72.6 CH | 4.09 (dd, 10.3, 3.6) | 123.1 CH | 7.36 (d, 7.2) |
| 8 | 76.2 C | 133.7 C | ||
| 8a | 117.0 C | 117.4 C | ||
| 9 | 182.0 C | 180.9 C | ||
| 9a | 109.6 C | 111.0 C | ||
| 10 | 167.4 C | 156.2 C | ||
| 11 | 65.0 CH2 | 5.13 s | 165.3 C | |
| 12 | 172.7 C | 169.2 C | ||
| 11-OMe | 52.8 CH3 | 3.98 s | ||
| 11-OAc | 170.5 C | |||
| 20.8 CH3 | 2.16 s | |||
| 12-OMe | 53.4 CH3 | 3.84 s | 53.2 CH3 | 4.04 s |
| 1-OH | 11.99 s | 12.20 s | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Y.-J.; Zou, Z.-B.; Xie, M.-M.; Zhang, Y.; Xu, L.; Yu, H.-Y.; Ma, H.-B.; Yang, X.-W. Ferroptosis Inhibitory Compounds from the Deep-Sea-Derived Fungus Penicillium sp. MCCC 3A00126. Mar. Drugs 2023, 21, 234. https://doi.org/10.3390/md21040234
Hao Y-J, Zou Z-B, Xie M-M, Zhang Y, Xu L, Yu H-Y, Ma H-B, Yang X-W. Ferroptosis Inhibitory Compounds from the Deep-Sea-Derived Fungus Penicillium sp. MCCC 3A00126. Marine Drugs. 2023; 21(4):234. https://doi.org/10.3390/md21040234
Chicago/Turabian StyleHao, You-Jia, Zheng-Biao Zou, Ming-Min Xie, Yong Zhang, Lin Xu, Hao-Yu Yu, Hua-Bin Ma, and Xian-Wen Yang. 2023. "Ferroptosis Inhibitory Compounds from the Deep-Sea-Derived Fungus Penicillium sp. MCCC 3A00126" Marine Drugs 21, no. 4: 234. https://doi.org/10.3390/md21040234