Hepialiamides A–C: Aminated Fusaric Acid Derivatives and Related Metabolites with Anti-Inflammatory Activity from the Deep-Sea-Derived Fungus Samsoniella hepiali W7
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Identification, Fermentation, and Extract
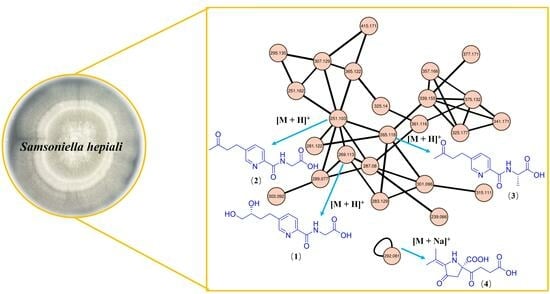
3.3. UHPLC-Q-TOF and Molecular Networking Analysis
3.4. Isolation and Purification
3.5. Theoretical ECD Calculation
3.6. BV-2 Cell Culture and Treatment
3.7. Nitrite Quantification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Haque, N.; Parveen, S.; Tang, T.; Wei, J.; Huang, Z. Marine natural products in clinical use. Mar. Drugs 2022, 20, 528. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.F.; Xu, L.; Wei, M.Y.; Wang, C.Y.; Gu, Y.C.; Shao, C.L. Recent progresses in marine microbial-derived antiviral natural products. Arch. Pharm. Res. 2020, 43, 1215–1229. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef]
- Hai, Y.; Cai, Z.M.; Li, P.J.; Wei, M.Y.; Wang, C.Y.; Gu, Y.C.; Shao, C.L. Trends of antimalarial marine natural products: Progresses, challenges and opportunities. Nat. Prod. Rep. 2022, 39, 969–990. [Google Scholar] [CrossRef]
- Hafez Ghoran, S.; Taktaz, F.; Ayatollahi, S.A.; Kijjoa, A. Anthraquinones and their analogues from marine-derived fungi: Chemistry and biological activities. Mar. Drugs 2022, 20, 474. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2022, 39, 1122–1171. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2023, 40, 275–325. [Google Scholar] [CrossRef]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; de Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.F.; Petras, D.; Schmid, R.; Duhrkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef]
- Wei, X.; Su, J.C.; Hu, J.S.; He, X.X.; Lin, S.J.; Zhang, D.M.; Ye, W.C.; Chen, M.F.; Lin, H.W.; Zhang, C.X. Probing indole diketopiperazine-based hybrids as environmental-induced products from Aspergillus sp. EGF 15-0-3. Org. Lett. 2022, 24, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.J.; Tian, D.M.; Chen, M.; Xia, Z.X.; Tang, X.Y.; Zhang, S.H.; Wei, J.H.; Li, X.N.; Yao, X.S.; Wu, B.; et al. Molecular networking-guided isolation of cyclopentapeptides from the hydrothermal vent sediment derived fungus Aspergillus pseudoviridinutans TW58-5 and their anti-inflammatory effects. J. Nat. Prod. 2023, 86, 1919–1930. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, J.; Hu, L.; Bai, R.; Wang, T.; Xing, X.; Chen, L.; Ding, G. LC-MS/MS-guided molecular networking for targeted discovery of undescribed and bioactive ophiobolins from Bipolaris eleusines. J. Agric. Food Chem. 2023, 71, 11982–11992. [Google Scholar] [CrossRef] [PubMed]
- Dobson, T.A.; Desaty, D.; Brewer, D.; Vining, L.C. Biosynthesis of fusaric acid in cultures of Fusarium oxysporum Schlecht. Can. J. Biochem. 1967, 45, 809–823. [Google Scholar] [CrossRef]
- Stipanovic, R.D.; Wheeler, M.H.; Puckhaber, L.S.; Liu, J.; Bell, A.A.; Williams, H.J. Nuclear magnetic resonance (NMR) studies on the biosynthesis of fusaric acid from Fusarium oxysporum f. sp. vasinfectum. J. Agric. Food Chem. 2011, 59, 5351–5356. [Google Scholar] [CrossRef] [PubMed]
- Studt, L.; Janevska, S.; Niehaus, E.M.; Burkhardt, I.; Arndt, B.; Sieber, C.M.; Humpf, H.U.; Dickschat, J.S.; Tudzynski, B. Two separate key enzymes and two pathway-specific transcription factors are involved in fusaric acid biosynthesis in Fusarium fujikuroi. Environ. Microbiol. 2016, 18, 936–956. [Google Scholar] [CrossRef] [PubMed]
- Hai, Y.; Chen, M.; Huang, A.; Tang, Y. Biosynthesis of mycotoxin fusaric acid and application of a PLP-dependent enzyme for chemoenzymatic synthesis of substituted l-pipecolic acids. J. Am. Chem. Soc. 2020, 142, 19668–19677. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Dan, W.J.; Li, Y.X.; Peng, G.R.; Zhang, A.L.; Gao, J.M. Antifungal metabolites from Alternaria atrans: An endophytic fungus in Psidium guajava. Nat. Prod. Commun. 2019, 14, 1934578X19844116. [Google Scholar] [CrossRef]
- Hilário, F.; Chapla, V.M.; Araujo, A.R.; Sano, P.T.; Bauab, T.M.; dos Santos, L.C. Antimicrobial screening of endophytic fungi isolated from the aerial parts of Paepalanthus chiquitensis (Eriocaulaceae) led to the isolation of secondary metabolites produced by Fusarium fujikuroi. J. Braz. Chem. Soc. 2016, 28, 1389–1395. [Google Scholar] [CrossRef]
- Zou, Z.B.; Chen, L.H.; Hu, M.Y.; Xu, L.; Hao, Y.J.; Yan, Q.X.; Wang, C.F.; Xie, C.L.; Yang, X.W. Cladosporioles A and B, two new indole derivatives from the deep-sea-derived fungus Cladosporium cladosporioides 170056. Chem. Biodivers. 2022, 19, e202200538. [Google Scholar] [CrossRef]
- Zou, Z.B.; Zhang, G.; Zhou, Y.Q.; Xie, C.L.; Xie, M.M.; Xu, L.; Hao, Y.J.; Luo, L.Z.; Zhang, X.K.; Yang, X.W.; et al. Chemical constituents of the deep-sea-derived Penicillium citreonigrum MCCC 3A00169 and their antiproliferative effects. Mar. Drugs 2022, 20, 736. [Google Scholar] [CrossRef] [PubMed]
- He, Z.H.; Xie, C.L.; Wu, T.; Yue, Y.T.; Wang, C.F.; Xu, L.; Xie, M.M.; Zhang, Y.; Hao, Y.J.; Xu, R.; et al. Tetracyclic steroids bearing a bicyclo [4.4.1] ring system as potent antiosteoporosis agents from the deep-sea-derived fungus Rhizopus sp. W23. J. Nat. Prod. 2023, 86, 157–165. [Google Scholar] [CrossRef]
- He, Z.H.; Xie, C.L.; Wu, T.; Zhang, Y.; Zou, Z.B.; Xie, M.M.; Xu, L.; Capon, R.J.; Xu, R.; Yang, X.W. Neotricitrinols A-C, unprecedented citrinin trimers with anti-osteoporosis activity from the deep-sea-derived Penicillium citrinum W23. Bioorg. Chem. 2023, 139, 106756. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.D.; Yue, J.M. A total synthesis of (+)- and (−)-dihydrokavain with a sonochemical blaise reaction as the key step. Eur. J. Org. Chem. 2005, 2005, 2575–2579. [Google Scholar] [CrossRef]
- Roberts, A.; Beaumont, C.; Manzarpour, A.; Mantle, P. Purpurolic acid: A new natural alkaloid from Claviceps purpurea (Fr.) Tul. Fungal Biol. 2016, 120, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; He, X.; Wu, G.; Liu, C.; Lu, C.; Gu, Q.; Che, Q.; Zhu, T.; Zhang, G.; Li, D. Aniline-tetramic acids from the deep-sea-derived fungus Cladosporium sphaerospermum L3P3 cultured with the HDAC inhibitor SAHA. J. Nat. Prod. 2018, 81, 1651–1657. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Park, H.J.; Han, E.S.; Park, D.K. Inhibitory effect on B16/F10 mouse melanoma cell and HT-29 human colon cancer cell proliferation and cordycepin content of the butanol extract of Paecilomyces militaris. J. Med. Plant Res. 2011, 5, 1066–1071. [Google Scholar]
- Domondon, D.L.; He, W.; De Kimpe, N.; Hofte, M.; Poppe, J. Beta-adenosine, a bioactive compound in grass chaff stimulating mushroom production. Phytochemistry 2004, 65, 181–187. [Google Scholar] [CrossRef]
- Qiu, W.; Wu, J.; Choi, J.H.; Hirai, H.; Nishida, H.; Kawagishi, H. Cytotoxic compounds against cancer cells from Bombyx mori inoculated with Cordyceps militaris. Biosci. Biotechnol. Biochem. 2017, 81, 1224–1226. [Google Scholar] [CrossRef]
- Tatani, K.; Hiratochi, M.; Nonaka, Y.; Isaji, M.; Shuto, S. Identification of 8-aminoadenosine derivatives as a new class of human concentrative nucleoside transporter 2 inhibitors. ACS. Med. Chem. Lett. 2015, 6, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Wu, L.; Ding, Y.; Cui, X.; Han, Z.; Xu, H. A new nucleoside and two new pyrrole alkaloid derivatives from Cordyceps militaris. Nat. Prod. Res. 2020, 34, 341–350. [Google Scholar] [CrossRef]
- Youssef, D.T.A.; Badr, J.M.; Shaala, L.A.; Mohamed, G.A.; Bamanie, F.H. Ehrenasterol and biemnic acid; new bioactive compounds from the Red Sea sponge Biemna ehrenbergi. Phytochem. Lett. 2015, 12, 296–301. [Google Scholar] [CrossRef]
- Ma, Y.T.; Qiao, L.R.; Shi, W.Q.; Zhang, A.L.; Gao, J.M. Metabolites produced by an endophyte Alternaria alternata isolated from Maytenus hookeri. Chem. Nat. Compd. 2010, 46, 504–506. [Google Scholar] [CrossRef]
- Ying, Y.M.; Shan, W.G.; Liu, W.H.; Zhan, Z.J. Alkaloids and nucleoside derivatives from a fungal endophyte of Huperzia serrata. Chem. Nat. Comd. 2013, 49, 184–186. [Google Scholar] [CrossRef]
- Kaaniche, F.; Hamed, A.; Abdel-Razek, A.S.; Wibberg, D.; Abdissa, N.; El Euch, I.Z.; Allouche, N.; Mellouli, L.; Shaaban, M.; Sewald, N. Bioactive secondary metabolites from new endophytic fungus Curvularia. sp isolated from Rauwolfia macrophylla. PLoS ONE 2019, 14, e0217627. [Google Scholar] [CrossRef]
- Sun, Y.; Tian, L.; Huang, J.; Li, W.; Pei, Y.H. Cytotoxic sterols from marine-derived fungus Pennicillium sp. Nat. Prod. Res. 2006, 20, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.T.; Wang, Y.J.; Ma, X.Y.; Yin, X.L.; Ji, N.Y. Two new sesquiterpenoids from the marine-sediment-derived fungus Trichoderma harzianum P1-4. Nat. Prod. Res. 2019, 33, 3127–3133. [Google Scholar] [CrossRef]
- Dearman, R.J.; Cumberbatch, M.; Hilton, J.; Clowes, H.M.; Fielding, I.; Heylings, J.R.; Kimber, I. Influence of dibutyl phthalate on dermal sensitization to fluorescein isothiocyanate. Fundam. Appl. Toxicol. 1996, 33, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Zeiger, E.; Haworth, S.; Mortelmans, K.; Speck, W. Mutagenicity testing of di(2-ethylhexyl)phthalate and related chemicals in Salmonella. Environ. Mutagen. 1985, 7, 213–232. [Google Scholar] [CrossRef]
- Wang, W.; Guo, J.; Zhang, J.; Liu, T.; Xin, Z. New screw lactam and two new carbohydrate derivatives from the methanol extract of rice bran. J. Agric. Food. Chem. 2014, 62, 10744–10751. [Google Scholar] [CrossRef]
- Theil, F.; Schick, H. Enzymes in organic-synthesis. 5. An improved procedure for the regioselective acetylation of monosaccharide derivatives by pancreatin-catalyzed transesterification in organic-solvents. Synthesis 1991, 22, 533–535. [Google Scholar] [CrossRef]
- Park, S.; Kazlauskas, R.J. Improved preparation and use of room-temperature ionic liquids in lipase-catalyzed enantio- and regioselective acylations. J. Org. Chem. 2001, 66, 8395–8401. [Google Scholar] [CrossRef] [PubMed]
- Li, X.B.; Li, Y.L.; Zhou, J.C.; Yuan, H.Q.; Wang, X.N.; Lou, H.X. A new diketopiperazine heterodimer from an endophytic fungus Aspergillus niger. J. Asian Nat. Prod. Res. 2015, 17, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.J.; Gao, S.S.; Li, C.S.; Li, X.M.; Wang, B.G. Chemical constituents of a marine-derived endophytic fungus Penicillium commune G2M. Molecules 2010, 15, 3270–3275. [Google Scholar] [CrossRef]
- Tao, Y.; Feng, C.; Xu, J.; Shen, L.; Qu, J.; Ju, H.; Yan, L.; Chen, W.; Zhang, Y. Di(2-ethylhexyl) phthalate and dibutyl phthalate have a negative competitive effect on the nitrification of black soil. Chemosphere 2022, 293, 133554. [Google Scholar] [CrossRef]
- Silvin, A.; Uderhardt, S.; Piot, C.; Da Mesquita, S.; Yang, K.; Geirsdottir, L.; Mulder, K.; Eyal, D.; Liu, Z.; Bridlance, C.; et al. Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity 2022, 55, 1448–1465. [Google Scholar] [CrossRef]
- Orihuela, R.; McPherson, C.A.; Harry, G.J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 2016, 173, 649–665. [Google Scholar] [CrossRef]





| No. | 1 a (δC, Type) | 1 a (δH, Mult J in Hz) | 2 b (δC, Type) | 2 b (δH, Mult J in Hz) | 3 a (δC, Type) | 3 a (δH, Mult J in Hz) |
|---|---|---|---|---|---|---|
| 2 | 147.3, C | 148.4, C | 147.4, C | |||
| 3 | 121.7, CH | 7.96, d (8.0) | 122.8, CH | 7.96, d (8.0) | 121.6, CH | 7.93, d (7.6) |
| 4 | 137.4, CH | 7.83, dd (2.0, 8.0) | 138.3, CH | 7.77, dd (2.0, 8.0) | 137.4, CH | 7.82, d (8.0) |
| 5 | 141.4, C | 141.9, C | 141.3, C | |||
| 6 | 148.5, CH | 8.51, d (1.5) | 150.1, CH | 8.47, d (1.6) | 148.7, CH | 8.51, s |
| 7 | 164.2, C | 167.0, C | 163.5, C | |||
| 8 | 28.3, CH2 | 2.71, m; 2.82, m | 27.5, CH2 | 2.90, d (5.2) | 26.1, CH2 | 2.85, m |
| 9 | 34.7, CH2 | 1.56, m; 1.77, m | 44.6, CH2 | 2.88, d (5.2) | 43.3, CH2 | 2.84, m |
| 10 | 70.2, CH | 3.39, m | 209.8, C | 207.3, C | ||
| 11 | 65.8, CH2 | 3.25, dd (5.7, 10.7) | 29.9, CH3 | 2.13, s | 29.7, CH3 | 2.09, s |
| 3.32, dd (5.6, 10.7) | ||||||
| 1′ | 41.0, CH2 | 3.97, s | 41.9, CH2 | 4.15, s | 47.6, CH | 4.46, d (7.1) |
| 2′ | 171.1, C | 172.8, C | 173.8, C | |||
| 3′ | 17.5, CH3 | 1.42, d (7.1) | ||||
| NH | 8.92, t (6.0) | 9.04, t (5.4) | 8.74, d (7.3) |
| No. | δC, Type | δH, Mult (J in Hz) |
|---|---|---|
| 2 | 70.3, C | |
| 3 | 47.4, CH2 | 3.15, d (18.0); 3.06, d (18.0) |
| 4 | 197.0, C | |
| 5 | 122.3, C | |
| 6 | 129.1, C | |
| 7 | 18.5, CH3 | 2.08, s |
| 8 | 20.8, CH3 | 1.81, s |
| 9 | 171.4, C | |
| 1′ | 207.2, C | |
| 2′ | 27.4, CH2 | 2.28, t (6.3) |
| 3′ | 36.3, CH2 | 2.68, m; 2.60, m |
| 4′ | 173.5, C | |
| NH | 10.27, s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, Z.-B.; Wu, T.-Z.; Yang, L.-H.; He, X.-W.; Liu, W.-Y.; Zhang, K.; Xie, C.-L.; Xie, M.-M.; Zhang, Y.; Yang, X.-W.; et al. Hepialiamides A–C: Aminated Fusaric Acid Derivatives and Related Metabolites with Anti-Inflammatory Activity from the Deep-Sea-Derived Fungus Samsoniella hepiali W7. Mar. Drugs 2023, 21, 596. https://doi.org/10.3390/md21110596
Zou Z-B, Wu T-Z, Yang L-H, He X-W, Liu W-Y, Zhang K, Xie C-L, Xie M-M, Zhang Y, Yang X-W, et al. Hepialiamides A–C: Aminated Fusaric Acid Derivatives and Related Metabolites with Anti-Inflammatory Activity from the Deep-Sea-Derived Fungus Samsoniella hepiali W7. Marine Drugs. 2023; 21(11):596. https://doi.org/10.3390/md21110596
Chicago/Turabian StyleZou, Zheng-Biao, Tai-Zong Wu, Long-He Yang, Xi-Wen He, Wen-Ya Liu, Kai Zhang, Chun-Lan Xie, Ming-Min Xie, Yong Zhang, Xian-Wen Yang, and et al. 2023. "Hepialiamides A–C: Aminated Fusaric Acid Derivatives and Related Metabolites with Anti-Inflammatory Activity from the Deep-Sea-Derived Fungus Samsoniella hepiali W7" Marine Drugs 21, no. 11: 596. https://doi.org/10.3390/md21110596