Extraction and Identification of Three New Urechis unicinctus Visceral Peptides and Their Antioxidant Activity
Abstract
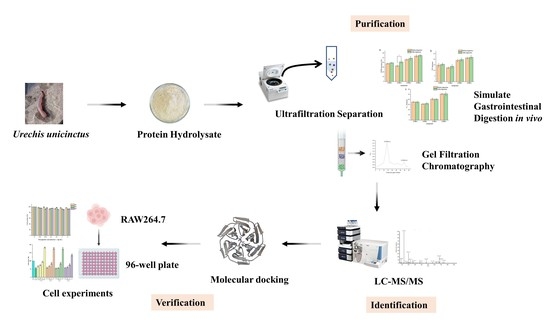
:1. Introduction
2. Results and Discussion
2.1. Degree of Hydrolysis (DH)
2.2. Ultrafiltration and Determination of Antioxidant Activity before and after Simulated Gastrointestinal Digestion
2.3. Gel-Filtration Chromatography
2.4. Identification of Peptide Sequences
2.5. The Structure–Activity Relationship
2.5.1. Induction
2.5.2. Molecular Docking
2.6. Cytoprotective Activity of Peptide on H2O2-Damaged RAW264.7 Cells
2.6.1. Cytotoxicity
2.6.2. Protection of Peptide on H2O2-Induced Oxidative Damage RAW264.7 Cells
3. Materials and Methods
3.1. Materials
3.2. Preparation of Protein Hydrolysate from Urechis unicinctus Viscera (UUPH)
3.3. Degree of Hydrolysis (DH)
3.4. Isolation and Purification of UUPH
3.4.1. Ultrafiltration Separation
3.4.2. Simulation of Gastrointestinal Digestion In Vivo
3.4.3. Gel Filtration Chromatography (GFC)
3.5. Peptide Identification by LC-MS/MS
3.6. Molecular Modeling
3.7. Antioxidant Activity of the Peptides
3.7.1. DPPH· Scavenging Activity
3.7.2. Hydroxyl Radicals (·OH) Scavenging Activity
3.7.3. Superoxide Anion Free Radical (·O2−) Scavenging Activity
3.8. Cell Cytotoxicity Assay
3.9. The Cytoprotective Activity of Antioxidant Peptide on Oxidative Damaged RW264.7 Cells by H2O2
3.10. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, X.; Liu, X.; Zhou, D.; Bai, Y.; Gao, B.; Zhang, Z.; Qin, Z. The NF-κB pathway participates in the response to sulfide stress in Urechis unicinctus. Fish Shellfish. Immunol. 2016, 58, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Tang, Y.Z.; Song, S.L.; Wang, B.G. Studies on technique for artificial breeding of Urechis unicinctus. Shandong Fish. 1997, 1, 5–8. [Google Scholar]
- Chen, W.; Zhang, S.; Sun, Y.; Tian, B.; Song, L.; Xu, Y.; Liu, T. Effects of substrate on the physiological characteristics and intestinal microbiota of Echiura worm (Urechis unicinctus) juveniles. Aquaculture 2021, 530, 735710. [Google Scholar] [CrossRef]
- Ryu, B.M.; Kim, M.J.; Himaya, S.W.A.; Kang, K.H.; Kim, S.K. Statistical optimization of high temperature/pressure and ultra-wave assisted lysis of Urechis unicinctus for the isolation of active peptide which enhance the erectile function in vitro. Process Biochem. 2014, 49, 148–153. [Google Scholar] [CrossRef]
- Kang, H.K.; Lee, H.H.; Seo, C.H.; Park, Y. Antimicrobial and Immunomodulatory Properties and Applications of Marine-Derived Proteins and Peptides. Mar. Drugs 2019, 17, 350. [Google Scholar] [CrossRef] [Green Version]
- Harnedy, P.A.; FitzGerald, R.J. Bioactive peptides from marine processing waste and shellfish: A review. J. Funct. Foods 2012, 4, 6–24. [Google Scholar] [CrossRef]
- Kim, K.S.; Bae, W.J.; Kim, S.J.; Kang, K.H.; Kim, S.K.; Cho, H.J.; Hong, S.H.; Lee, J.Y.; Kim, S.W. Improvement of erectile dysfunction by the active pepide from Urechis unicinctus by high temperature/pressure and ultra—Wave assisted lysis in Streptozotocin Induced Diabetic Rats. Int. Braz. J. Urol. 2016, 42, 825–837. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Ma, Y.; Zuo, Y.; Liu, Z.; Wang, Q.; Ren, D.; He, Y.; Cong, H.; Wu, L.; Zhou, H. The efficient enrichment of marine peptides from the protein hydrolysate of the marine worm Urechis unicinctus by using mesoporous materials MCM-41, SBA-15 and CMK-3. Anal. Methods 2021, 13, 2405–2414. [Google Scholar] [CrossRef]
- Chen, M.L.; Ning, P.; Jiao, Y.; Xu, Z.; Cheng, Y.H. Extraction of antioxidant peptides from rice dreg protein hydrolysate via an angling method. Food Chem. 2021, 337, 128069. [Google Scholar] [CrossRef]
- Yang, Q.; Cai, X.; Yan, A.; Tian, Y.; Du, M.; Wang, S. A specific antioxidant peptide: Its properties in controlling oxidation and possible action mechanism. Food Chem. 2020, 327, 126984. [Google Scholar] [CrossRef]
- Haider, K.; Haider, M.R.; Neha, K.; Yar, M.S. Free radical scavengers: An overview on heterocyclic advances and medicinal prospects. Eur. J. Med. Chem. 2020, 204, 112607. [Google Scholar] [CrossRef] [PubMed]
- Dao, V.T.; Casas, A.I.; Maghzal, G.J.; Seredenina, T.; Kaludercic, N.; Robledinos-Anton, N.; Di Lisa, F.; Stocker, R.; Ghezzi, P.; Jaquet, V.; et al. Pharmacology and Clinical Drug Candidates in Redox Medicine. Antioxid. Redox. Signal. 2015, 23, 1113–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhia, S.; Kumar, A.; Kumar, R. Generation of antioxidant peptides from soy protein isolate through psychrotrophic Chryseobacterium sp. derived alkaline broad temperature active protease. LWT 2021, 143, 111152. [Google Scholar] [CrossRef]
- Torres-Fuentes, C.; Alaiz, M.; Vioque, J. Iron-chelating activity of chickpea protein hydrolysate peptides. Food Chem. 2012, 134, 1585–1588. [Google Scholar] [CrossRef]
- Zhang, X.X.; Liu, F.; Liu, C.E.; Ji, Y.; Duan, D.; Yan, N. Optimization of enzymatic hydrolysis of visceral polypeptides of echinus unicyclicus by Response Surface Methodology. Food Res. Dev. 2018, 39, 52–57. [Google Scholar]
- Jorge, S.; Pereira, K.; López-Fernández, H.; LaFramboise, W.; Dhir, R.; Fernández-Lodeiro, J.; Lodeiro, C.; Santos, H.M.; Capelo-Martínez, J.L. Ultrasonic-assisted extraction and digestion of proteins from solid biopsies followed by peptide sequential extraction hyphenated to MALDI-based profiling holds the promise of distinguishing renal oncocytoma from chromophobe renal cell carcinoma. Talanta 2020, 206, 120180. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Y.M.; Chi, C.F.; Luo, H.Y.; Deng, S.G.; Ma, J.Y. Isolation and characterization of collagen and antioxidant collagen peptides from scales of croceine croaker (Pseudosciaena crocea). Mar. Drugs 2013, 11, 4641–4661. [Google Scholar] [CrossRef]
- Xing, L.; Liu, R.; Gao, X.; Zheng, J.; Wang, C.; Zhou, G.; Zhang, W. The proteomics homology of antioxidant peptides extracted from dry-cured Xuanwei and Jinhua ham. Food Chem. 2018, 266, 420–426. [Google Scholar] [CrossRef]
- Canabady-Rochelle, L.L.; Harscoat-Schiavo, C.; Kessler, V.; Aymes, A.; Fournier, F.; Girardet, J.M. Determination of reducing power and metal chelating ability of antioxidant peptides: Revisited methods. Food Chem. 2015, 183, 129–135. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Fu, X.; Li, S.; Wei, J. A novel antioxidant and ACE inhibitory peptide from rice bran protein: Biochemical characterization and molecular docking study. LWT 2017, 75, 93–99. [Google Scholar] [CrossRef]
- Moayedi, A.; Mora, L.; Aristoy, M.C.; Safari, M.; Hashemi, M.; Toldrá, F. Peptidomic analysis of antioxidant and ACE-inhibitory peptides obtained from tomato waste proteins fermented using Bacillus subtilis. Food Chem. 2018, 250, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.X.; Xu, H.P.; Li, Y.; Zhang, Q.W.; Xie, H. Preparation and Evaluation of Peptides with Potential Antioxidant Activity by Microwave Assisted Enzymatic Hydrolysis of Collagen from Sea Cucumber Acaudina Molpadioides Obtained from Zhejiang Province in China. Mar. Drugs 2019, 17, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, F.; Ci, A.T.; Wang, H.; Zhang, Y.Y.; Zhang, J.G.; Thakur, K.; Wei, Z.J. Identification and hydrolysis kinetic of a novel antioxidant peptide from pecan meal using Alcalase. Food Chem. 2018, 261, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Zhang, J.; Zhang, H.; Duan, Y.; Ma, H. Plant protein-derived antioxidant peptides: Isolation, identification, mechanism of action and application in food systems: A review. Trends Food Sci. Technol. 2020, 105, 308–322. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, L.; Sun, Q.; Song, G.; Huang, J. Extraction, identification and structure-activity relationship of antioxidant peptides from sesame (Sesamum indicum L.) protein hydrolysate. Food Res. Int. 2019, 116, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wu, Y.; Li, L. Relationship between primary structure or spatial conformation and functional activity of antioxidant peptides from Pinctada fucata. Food Chem. 2018, 264, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Li, M.; Ding, J.; Zheng, J.; Zhu, B.W.; Lin, S. Structure-activity relationship and pathway of antioxidant shrimp peptides in a PC12 cell model. J. Funct. Foods 2020, 70, 103978. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, S.; Wu, D.; Xu, S.; Chen, H.; Du, M. Hydrophobic peptides from oyster protein hydrolysates show better zinc-chelating ability. Food Biosci. 2021, 41, 100985. [Google Scholar] [CrossRef]
- Tonolo, F.; Folda, A.; Cesaro, L.; Scalcon, V.; Marin, O.; Ferro, S.; Bindoli, A.; Rigobello, M.P. Milk-derived bioactive peptides exhibit antioxidant activity through the Keap1-Nrf2 signaling pathway. J. Funct. Foods 2020, 64, 103696. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Lin, S.J.; Yang, Z.S.; Jin, H.X. Preparation of Antioxidant Peptide by Microwave- Assisted Hydrolysis of Collagen and Its Protective Effect Against H2O2-Induced Damage of RAW264.7 Cells. Mar. Drugs 2019, 17, 642. [Google Scholar] [CrossRef] [Green Version]
- GB5009.235-2016; State Health and Family Planning Commission of the People’s Republic of China. National Standard of the People’s Republic of China–Determination of Amino Acid Nitrogen in Food. Standards Press: Beijing, China, 2016.
- Song, Y.; Fu, Y.; Huang, S.; Liao, L.; Wu, Q.; Wang, Y.; Ge, F.; Fang, B. Identification and antioxidant activity of bovine bone collagen-derived novel peptides prepared by recombinant collagenase from Bacillus cereus. Food Chem. 2021, 349, 129143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, M.; Zhang, G.; Tian, Y.; Kong, F.; Xiong, S.; Zhao, S.; Jia, D.; Manyande, A.; Du, H. Identification of novel antioxidant peptides from snakehead (Channa argus) soup generated during gastrointestinal digestion and insights into the anti-oxidation mechanisms. Food Chem. 2021, 337, 127921. [Google Scholar] [CrossRef] [PubMed]








| No. | RT (min) | Amino Acid Sequence | MW (Da) |
|---|---|---|---|
| P1 | 31.52 | VTSALVGPR | 898.5 |
| P2 | 43.87 | IGLGDEGLRR | 1084.6 |
| P3 | 56.72 | TKIRNEISDLNER | 1586.8 |
| Amino Acids | P1 | P2 | P3 |
|---|---|---|---|
| A(Ala) | 8.84 | 0 | 0 |
| G(Gly) | 7.2 | 18.05 | 0 |
| L(Leu) | 12.57 | 21.03 | 7.27 |
| P(Pro) | 11.04 | 0 | 0 |
| V(Val) | 22.46 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Lu, J.; Asakiya, C.; Huang, K.; Zhou, X.; Liu, Q.; He, X. Extraction and Identification of Three New Urechis unicinctus Visceral Peptides and Their Antioxidant Activity. Mar. Drugs 2022, 20, 293. https://doi.org/10.3390/md20050293
Li J, Lu J, Asakiya C, Huang K, Zhou X, Liu Q, He X. Extraction and Identification of Three New Urechis unicinctus Visceral Peptides and Their Antioxidant Activity. Marine Drugs. 2022; 20(5):293. https://doi.org/10.3390/md20050293
Chicago/Turabian StyleLi, Jingjing, Jiajun Lu, Charles Asakiya, Kunlun Huang, Xiuzhi Zhou, Qingliang Liu, and Xiaoyun He. 2022. "Extraction and Identification of Three New Urechis unicinctus Visceral Peptides and Their Antioxidant Activity" Marine Drugs 20, no. 5: 293. https://doi.org/10.3390/md20050293