5-O-(N-Boc-l-Alanine)-Renieramycin T Induces Cancer Stem Cell Apoptosis via Targeting Akt Signaling
Abstract
:1. Introduction
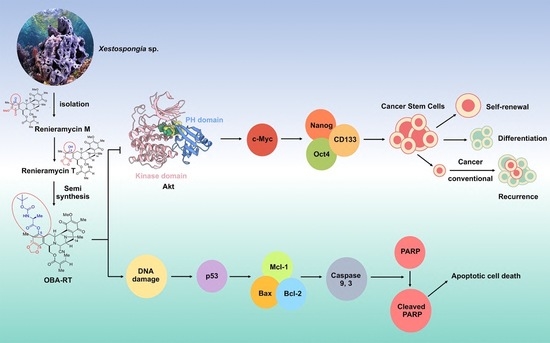
2. Results
2.1. Semi-Synthesis of 5-O-(N-Boc-l-Alanine)-Renieramycin T (OBA-RT)
2.2. Cytotoxicity and Apoptosis-Inducing Effect of OBA-RT
2.3. OBA-RT Induced Apoptosis through p53 Activation
2.4. OBA-RT Suppresses CSC Spheroid Formation
2.5. OBA-RT Suppresses CSC Signals in A549 Cells
2.6. Molecular Docking Simulations Indicated the OBA-RT Interactions with the Allosteric Pocket of Akt-1 Protein
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Semi-Synthesis of 5-O-(N-Boc-l-Alanine)-Renieramycin T (OBA-RT)
4.3. Preparation of the OBA-RT Stock Solution
4.4. Cell Lines and Culture
4.5. Cell Viability
4.6. Colony Formation Assay
4.7. Apoptotic Assay
4.8. Spheroid Formation Assay
4.9. CSC-Rich Population
4.10. Western Blot Analysis
4.11. Immunofluorescence Assay
4.12. Molecular Docking
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, Z.; Wang, Q.; Shigdar, S.; Duan, W.; Xiang, D. Cancer Stem Cells—Perspectives and How to Target Them. In Smart Drug Delivery System; IntechOpen: London, UK, 2016; pp. 117–137. [Google Scholar]
- Pan, Y.; Ma, S.; Cao, K.; Zhou, S.; Zhao, A.; Li, M.; Qian, F.; Zhu, C. Therapeutic approaches targeting cancer stem cells. J. Cancer Res. Ther. 2018, 14, 1469–1475. [Google Scholar] [CrossRef] [PubMed]
- Dragu, D.L.; Necula, L.G.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Therapies targeting cancer stem cells: Current trends and future challenges. World J. Stem Cells 2015, 7, 1185–1201. [Google Scholar] [CrossRef] [PubMed]
- Casey, S.C.; Baylot, V.; Felsher, D.W. The MYC oncogene is a global regulator of the immune response. Blood 2018, 131, 2007–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanvorachote, P.; Sriratanasak, N.; Nonpanya, N. C-myc Contributes to Malignancy of Lung Cancer: A Potential Anticancer Drug Target. Anticancer Res. 2020, 40, 609–618. [Google Scholar] [CrossRef]
- Zhang, H.F.; Wu, C.; Alshareef, A.; Gupta, N.; Zhao, Q.; Xu, X.E.; Jiao, J.W.; Li, E.M.; Xu, L.Y.; Lai, R. The PI3K/AKT/c-MYC Axis Promotes the Acquisition of Cancer Stem-Like Features in Esophageal Squamous Cell Carcinoma. Stem Cells 2016, 34, 2040–2051. [Google Scholar] [CrossRef] [Green Version]
- Tan, A.C. Targeting the PI3K/Akt/mTOR pathway in non-small cell lung cancer (NSCLC). Thorac. Cancer 2020, 11, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Demetri, G.D.; Von Mehren, M.; Jones, R.L.; Hensley, M.L.; Schuetze, S.M.; Staddon, A.; Milhem, M.; Elias, A.; Ganjoo, K.; Tawbi, H. Efficacy and safety of trabectedin or dacarbazine for metastatic liposarcoma or leiomyosarcoma after failure of conventional chemotherapy: Results of a phase III randomized multicenter clinical trial. J. Clin. Oncol. 2016, 34, 786. [Google Scholar] [CrossRef]
- Yokoi, E.; Mabuchi, S.; Shimura, K.; Komura, N.; Kozasa, K.; Kuroda, H.; Takahashi, R.; Sasano, T.; Kawano, M.; Matsumoto, Y. Lurbinectedin (PM01183), a selective inhibitor of active transcription, effectively eliminates both cancer cells and cancer stem cells in preclinical models of uterine cervical cancer. Investig. New Drugs 2019, 37, 818–827. [Google Scholar] [CrossRef]
- Hoda, M.A.; Pirker, C.; Dong, Y.; Schelch, K.; Heffeter, P.; Kryeziu, K.; van Schoonhoven, S.; Klikovits, T.; Laszlo, V.; Rozsas, A. Trabectedin is active against malignant pleural mesothelioma cell and xenograft models and synergizes with chemotherapy and Bcl-2 inhibition in vitro. Mol. Cancer Ther. 2016, 15, 2357–2369. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Serra, J.; Maffiotte, E.; Martín, J.; Bex, T.; Navarro-Palou, M.; Ros, T.; Plazas, J.M.; Vögler, O.; Gutiérrez, A.; Amat, J.C. Yondelis®(ET-743, Trabectedin) sensitizes cancer cell lines to CD95-mediated cell death: New molecular insight into the mechanism of action. Eur. J. Pharmacol. 2011, 658, 57–64. [Google Scholar] [CrossRef]
- Powan, P.; Saito, N.; Suwanborirux, K.; Chanvorachote, P. Ecteinascidin 770, a tetrahydroisoquinoline alkaloid, sensitizes human lung cancer cells to anoikis. Anticancer Res. 2013, 33, 505–512. [Google Scholar] [PubMed]
- Scott, J.D.; Williams, R.M. Chemistry and biology of the tetrahydroisoquinoline antitumor antibiotics. Chem. Rev. 2002, 102, 1669–1730. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Li, H.; Ji, B.; Cheng, K.; Wu, B.; Li, Z.; Zheng, C.; Hua, H.; Li, D. Renieramycin-type alkaloids from marine-derived organisms: Synthetic chemistry, biological activity and structural modification. Eur. J. Med. Chem. 2021, 210, 113092. [Google Scholar] [CrossRef] [PubMed]
- Sirimangkalakitti, N.; Chamni, S.; Suwanborirux, K.; Chanvorachote, P. Renieramycin M sensitizes anoikis-resistant H460 lung cancer cells to anoikis. Anticancer Res. 2016, 36, 1665–1671. [Google Scholar]
- Tatsukawa, M.; Punzalan, L.L.C.; Magpantay, H.D.; Villasenor, I.M.; Concepcion, G.P.; Suwanborirux, K.; Yokoya, M.; Saito, N. Chemistry of renieramycins. Part 13: Isolation and structure of stabilized renieramycin type derivatives, renieramycins W–Y, from Philippine blue sponge Xestospongia sp., pretreated with potassium cyanide. Tetrahedron 2012, 68, 7422–7428. [Google Scholar] [CrossRef]
- Sirimangkalakitti, N.; Chamni, S.; Suwanborirux, K.; Chanvorachote, P. Renieramycin m attenuates cancer stem cell-like phenotypes in h460 lung cancer cells. Anticancer Res. 2017, 37, 615–621. [Google Scholar] [CrossRef] [Green Version]
- Martinez, E.J.; Owa, T.; Schreiber, S.L.; Corey, E. Phthalascidin, a synthetic antitumor agent with potency and mode of action comparable to ecteinascidin 743. Proc. Natl. Acad. Sci. USA 1999, 96, 3496–3501. [Google Scholar] [CrossRef] [Green Version]
- Chamni, S.; Sirimangkalakitti, N.; Chanvorachote, P.; Suwanborirux, K.; Saito, N. Chemistry of renieramycins. Part 19: Semi-syntheses of 22-O-amino ester and hydroquinone 5-O-amino ester derivatives of renieramycin M and their cytotoxicity against non-small-cell lung cancer cell lines. Mar. Drugs 2020, 18, 418. [Google Scholar] [CrossRef]
- Maiuthed, A.; Pinkhien, T.; Chamni, S.; Suwanborirux, K.; Saito, N.; Petpiroon, N.; Chanvorachote, P. Apoptosis-inducing effect of hydroquinone 5-O-cinnamoyl ester analog of renieramycin M on non-small cell lung cancer cells. Anticancer. Res. 2017, 37, 6259–6267. [Google Scholar]
- Hongwiangchan, N.; Sriratanasak, N.; Wichadakul, D.; Aksorn, N.; Chamni, S.; Chanvorachote, P. Hydroquinone 5-O-Cinnamoyl Ester of Renieramycin M Suppresses Lung Cancer Stem Cells by Targeting Akt and Destabilizes c-Myc. Pharmaceuticals 2021, 14, 1112. [Google Scholar] [CrossRef]
- Daikuhara, N.; Tada, Y.; Yamaki, S.; Charupant, K.; Amnuoypol, S.; Suwanborirux, K.; Saito, N. Chemistry of renieramycins. Part 7: Renieramycins T and U, novel renieramycin–ecteinascidin hybrid marine natural products from Thai sponge Xestospongia sp. Tetrahedron Lett. 2009, 50, 4276–4278. [Google Scholar] [CrossRef]
- Petsri, K.; Chamni, S.; Suwanborirux, K.; Saito, N.; Chanvorachote, P. Renieramycin T Induces Lung Cancer Cell Apoptosis by Targeting Mcl-1 Degradation: A New Insight in the Mechanism of Action. Mar. Drugs 2019, 17, 301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chantarawong, W.; Chamni, S.; Suwanborirux, K.; Saito, N.; Chanvorachote, P. 5-O-Acetyl-Renieramycin T from Blue Sponge Xestospongia sp. Induces Lung Cancer Stem Cell Apoptosis. Mar. Drugs 2019, 17, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoya, M.; Toyoshima, R.; Suzuki, T.; Le, V.H.; Williams, R.M.; Saito, N. Stereoselective total synthesis of (−)-renieramycin T. J. Org. Chem. 2016, 81, 4039–4047. [Google Scholar] [CrossRef]
- He, W.; Zhang, Z.; Ma, D. A Scalable Total Synthesis of the Antitumor Agents Et-743 and Lurbinectedin. Angew. Chem. Int. Ed. 2019, 58, 3972–3975. [Google Scholar] [CrossRef] [PubMed]
- Cleves, A.; Jain, A. Knowledge-guided docking: Accurate prospective prediction of bound configurations of novel ligands using Surflex-Dock. J. Comput.-Aided Mol. Des. 2015, 29, 485–509. [Google Scholar] [PubMed] [Green Version]
- Ando, Y.; Suzuki, K. Photoredox reactions of quinones. Chem.–Eur. J. 2018, 24, 15955–15964. [Google Scholar] [CrossRef]
- Andrew, N.; Stephen, P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar]
- Nakajima, W.; Tanaka, N. The anti-apoptotic protein MCL1, a novel target of lung cancer therapy. J. Cancer Treat. Diagn. 2018, 2, 54–58. [Google Scholar]
- Yu, J.S.; Cui, W. Proliferation, survival and metabolism: The role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development 2016, 143, 3050–3060. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Yang, Y.; Li, W.; Chen, Q.; Li, J.; Pan, X.; Zhou, L.; Liu, C.; Chen, C.; He, J.; et al. Reciprocal regulation of Akt and Oct4 promotes the self-renewal and survival of embryonal carcinoma cells. Mol. Cell 2012, 48, 627–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, C.; Lu, J.; Yi, B.C.; Chang, K.K.; Simon, M.C.; Ryeom, S.; Yoon, S.S. PI3K/Akt pathway and Nanog maintain cancer stem cells in sarcomas. Oncogenesis 2021, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, N.; Heerding, D.A.; Duckett, D.R.; Eberwein, D.J.; Knick, V.B.; Lansing, T.J.; McConnell, R.T.; Gilmer, T.M.; Zhang, S.Y.; Robell, K.; et al. Characterization of an Akt kinase inhibitor with potent pharmacodynamic and antitumor activity. Cancer Res. 2008, 68, 2366–2374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, A.; Yu, X.; Liu, S. Pluripotency transcription factors and cancer stem cells: Small genes make a big difference. Chin. J. Cancer 2013, 32, 483. [Google Scholar] [CrossRef]
- Liu, G.; Shi, A.; Wang, N.; Li, M.; He, X.; Yin, C.; Tu, Q.; Shen, X.; Tao, Y.; Wang, Q. Polyphenolic Proanthocyanidin-B2 suppresses proliferation of liver cancer cells and hepatocellular carcinogenesis through directly binding and inhibiting AKT activity. Redox Biol. 2020, 37, 101701. [Google Scholar] [CrossRef]
- Yilmaz, O.G.; Olmez, E.O.; Ulgen, K.O. Targeting the Akt1 allosteric site to identify novel scaffolds through virtual screening. Comput. Biol. Chem. 2014, 48, 1–13. [Google Scholar] [CrossRef]
- Wu, W.I.; Voegtli, W.C.; Sturgis, H.L.; Dizon, F.P.; Vigers, G.P.; Brandhuber, B.J. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLoS ONE 2010, 5, e12913. [Google Scholar] [CrossRef]
- Yokoya, M.; Takahashi, S.; Saito, N. Preparation of Tricyclic Analog As Cde Ring Model of Renieramycin Marine Natural Product by Novel Photo-Induced Transformation of 6-Methoxy-1,2,3,4-Tetrahydroisoquinoline-5,8-Dione (Dedicated to Professor Tohru Fukuyama on the occasion of his 70th birthday). Heterocycles Int. J. Rev. Commun. Heterocycl. Chem. 2019, 99, 1276–1303. [Google Scholar]
- Lapierre, J.-M.; Eathiraj, S.; Vensel, D.; Liu, Y.; Bull, C.O.; Cornell-Kennon, S.; Iimura, S.; Kelleher, E.W.; Kizer, D.E.; Koerner, S. Discovery of 3-(3-(4-(1-Aminocyclobutyl)phenyl)-5-phenyl-3H-imidazo[4,5-b]pyridin-2-yl) pyridin-2-amine (ARQ 092): An orally bioavailable, selective, and potent allosteric AKT inhibitor. J. Med. Chem. 2016, 59, 6455–6469. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frisch, A. Gaussian 09W Reference; Gaussian, Inc.: Wallingford, CT, USA, 2009; 25p. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Compounds | Binding Energy (kcal/mol) | Hydrogen Bond Interactions | Hydrophobic Interactions |
|---|---|---|---|
| Miransertib (Co-crystal ligand) | −12.8 | Thr211, Tyr272 | Asn53, Gln79, Trp80, Thr82, Leu210, Leu264, Val270, Tyr272, Asp274, Ile290 |
| OBA-RT | −8.1 | Thr82, Glu203 | Asn53, Asn54, Ser56, Ala58, Gln79, Trp80, Leu202, Ser205, Leu264, Lys268, Val270, Asp292 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suksamai, D.; Racha, S.; Sriratanasak, N.; Chaotham, C.; Aphicho, K.; Lin, A.C.K.; Chansriniyom, C.; Suwanborirux, K.; Chamni, S.; Chanvorachote, P. 5-O-(N-Boc-l-Alanine)-Renieramycin T Induces Cancer Stem Cell Apoptosis via Targeting Akt Signaling. Mar. Drugs 2022, 20, 235. https://doi.org/10.3390/md20040235
Suksamai D, Racha S, Sriratanasak N, Chaotham C, Aphicho K, Lin ACK, Chansriniyom C, Suwanborirux K, Chamni S, Chanvorachote P. 5-O-(N-Boc-l-Alanine)-Renieramycin T Induces Cancer Stem Cell Apoptosis via Targeting Akt Signaling. Marine Drugs. 2022; 20(4):235. https://doi.org/10.3390/md20040235
Chicago/Turabian StyleSuksamai, Darinthip, Satapat Racha, Nicharat Sriratanasak, Chatchai Chaotham, Kanokpol Aphicho, Aye Chan Khine Lin, Chaisak Chansriniyom, Khanit Suwanborirux, Supakarn Chamni, and Pithi Chanvorachote. 2022. "5-O-(N-Boc-l-Alanine)-Renieramycin T Induces Cancer Stem Cell Apoptosis via Targeting Akt Signaling" Marine Drugs 20, no. 4: 235. https://doi.org/10.3390/md20040235