Seaweed Extracts: A Promising Source of Antibiofilm Agents with Distinct Mechanisms of Action against Pseudomonas aeruginosa
Abstract
:1. Introduction
2. Results
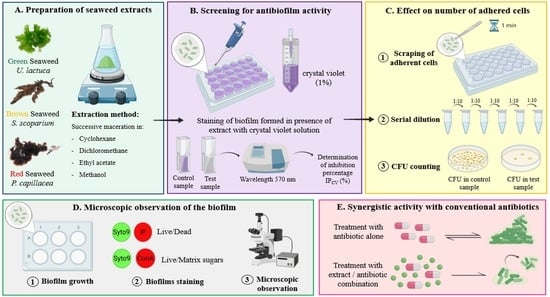
2.1. Extraction Yields of Different Seaweed Extracts
2.2. Assessment of the Inhibitory Effect of Extract on BIOFILM formation—Extracts Added at t0
2.2.1. Screening of Algal Extracts for Their Inhibitory Effect on PAO1 BIOFILM Formation and Growth—Crystal Violet (CV) Staining Method
2.2.2. Effect of Selected Active Extracts on the Number of Adhered Bacteria—CFU Counts Method
2.2.3. Phenotypic Observations of Biofilms by Epifluorescence Microscopy
2.3. Effect of Selected Algal Extracts on PAO1 24 h-Old Biofilm—Extracts Added at 24 h
2.4. Evaluation of the Synergistic Antibiofilm Activity of EA Extract in Combination with Tobramycin or Colistin
2.5. Analysis of the Chemical Composition of Extracts by GC–MS
3. Discussion
4. Materials and Methods
4.1. Collection of Algal Materials
4.2. Organic Solvents, Chemicals and Antibiotics
4.3. Bacterial Strain and Culture Media
4.4. Preparation of Seaweed Extracts
4.5. Assessment of the Inhibitory Effect of Extract on Biofilm Formation—Extract Added at t0
4.5.1. Formation of PAO1 Biofilms
4.5.2. Screening of Algal Extracts for Their Effect on PAO1 Biofilm Formation and Growth—Crystal Violet Staining Method
4.5.3. Effect of the Potentially Active Extracts on the Number of Adhered Bacteria—CFU Counts Method
4.5.4. Phenotypic Observations by Epifluorescence Microscopy
4.6. Effect of Selected Algal Extracts on PAO1 24 h-Old Biofilms—Extract Added at t24 h
4.7. Evaluation of the Synergistic Antibiofilm Activity of the Active Extract in Combination with Tobramycin or Colistin on 24 h-Old Treated Biofilms
4.8. Analysis of the Chemical Composition of Extracts by GC–MS
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Paul, D.; Gopal, J.; Kumar, M.; Manikandan, M. Nature to the natural rescue: Silencing microbial chats. Chem.-Biol. Interact. 2018, 280, 86–98. [Google Scholar] [CrossRef]
- Woolhouse, M.; Farrar, J. Policy: An intergovernmental panel on antimicrobial resistance. Nature 2014, 509, 555–557. [Google Scholar] [CrossRef] [Green Version]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Develipment of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, F.; Pham, D.T.N.; Tabassum, N.; Oloketuyi, S.F.; Kim, Y.M. Treatment strategies targeting persister cell formation in bacterial pathogens. Crit. Rev. Microbiol. 2020, 46, 665–688. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Campanac, C.; Pineau, L.; Payard, A.; Baziard-Mouysset, G.; Roques, C. Interactions between Biocide Cationic Agents and Bacterial Biofilms. Antimicrob. Agents Chemother. 2002, 46, 1469–1474. [Google Scholar] [CrossRef] [Green Version]
- Olivares, E.; Badel-Berchoux, S.; Provot, C.; Prévost, G.; Bernardi, T.; Jehl, F. Clinical Impact of Antibiotics for the Treatment of Pseudomonas aeruginosa Biofilm Infections. Front. Microbiol. 2020, 10, 2894. [Google Scholar] [CrossRef]
- Talebi Bezmin Abadi, A.; Rizvanov, A.A.; Haertlé, T.; Blatt, N.L. World Health Organization Report: Current Crisis of Antibiotic Resistance. BioNanoScience 2019, 9, 778–788. [Google Scholar] [CrossRef]
- Mishra, R.; Panda, A.K.; De Mandal, S.; Shakeel, M.; Bisht, S.S.; Khan, J. Natural Anti-biofilm Agents: Strategies to Control Biofilm-Forming Pathogens. Front. Microbiol. 2020, 11, 566325. [Google Scholar] [CrossRef]
- Dahms, H.; Dobretsov, S. Antifouling Compounds from Marine Macroalgae. Mar. Drugs 2017, 15, 265. [Google Scholar] [CrossRef] [PubMed]
- Bamunuarachchi, N.I.; Khan, F.; Kim, Y.M. Bactericidal activity of Sargassum aquifolium (Turner) C. Agardh against Gram-positive and Gram-negative biofilm-forming pathogenic bacteria. Curr. Pharm. Biotechnol. 2021, 22, 1628–1640. [Google Scholar] [CrossRef] [PubMed]
- Leandro, A.; Pereira, L.; Goncalves, A.M.M. Diverse Applications of Marine Macroalgae. Mar. Drugs 2019, 18, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosa, G.P.; Tavares, W.R.; Sousa, P.M.C.; Pagès, A.K.; Seca, A.M.L.; Pinto, D.C.G.A. Seaweed Secondary Metabolites with Beneficial Health Effects: An Overview of Successes in In Vivo Studies and Clinical Trials. Mar. Drugs 2019, 18, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, A.; Silva, S.A.; Carpena, M.; Garcia-Oliveira, P.; Gullón, P.; Barroso, M.F.; Prieto, M.A.; Simal-Gandara, J. Macroalgae as a Source of Valuable Antimicrobial Compounds: Extraction and Applications. Antibiotics 2020, 9, 642. [Google Scholar] [CrossRef]
- Manefield, M.; de Nys, R.; Naresh, K.; Roger, R.; Givskov, M.; Peter, S.; Kjelleberg, S. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 1999, 145, 283–291. [Google Scholar] [CrossRef] [Green Version]
- Guzzo, F.; Scognamiglio, M.; Fiorentino, A.; Buommino, E.; D’Abrosca, B. Plant Derived Natural Products against Pseudomonas aeruginosa and Staphylococcus aureus: Antibiofilm Activity and Molecular Mechanisms. Molecules 2020, 25, 5024. [Google Scholar] [CrossRef]
- Kim, H.-S.; Lee, S.-H.; Byun, Y.; Park, H.-D. 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Sci. Rep. 2015, 5, 8656. [Google Scholar] [CrossRef]
- Bhowmick, S.; Mazumdar, A.; Moulick, A.; Adam, V. Algal metabolites: An inevitable substitute for antibiotics. Biotechnol. Adv. 2020, 43, 107571. [Google Scholar] [CrossRef]
- Tang, J.; Wang, W.; Chu, W. Antimicrobial and Anti-Quorum Sensing Activities of Phlorotannins From Seaweed (Hizikia fusiforme). Front. Cell Infect. Microbiol. 2020, 10, 586750. [Google Scholar] [CrossRef]
- Salem, D.M.S.A.; Ismail, M.M.; Tadros, H.R.Z. Evaluation of the antibiofilm activity of three seaweed species and their biosynthesized iron oxide nanoparticles (Fe3O4-NPs). Egypt. J. Aquat. Res. 2020, 46, 333–339. [Google Scholar] [CrossRef]
- Cabral, E.M.; Oliveira, M.; Mondala, J.R.M.; Curtin, J.; Tiwari, B.K.; Garcia-Vaquero, M. Antimicrobials from Seaweeds for Food Applications. Mar. Drugs 2021, 19, 211. [Google Scholar] [CrossRef]
- Barreto, M.; Meyer, J.J.M. Isolation and antimicrobial activity of a lanosol derivative from Osmundaria serrata (Rhodophyta) and a visual exploration of its biofilm covering. S. Afr. J. Bot. 2006, 72, 521–528. [Google Scholar] [CrossRef] [Green Version]
- Tappenden, P.; Harnan, S.; Uttley, L.; Mildred, M.; Carroll, C.; Cantrell, A. Colistimethate sodium powder and tobramycin powder for inhalation for the treatment of chronic Pseudomonas aeruginosa lung infection in cystic fibrosis: Systematic review and economic model. Health Technol. Assess. 2013, 17, 1–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalilzadeh, P.; Lajoie, B.; El Hage, S.; Furiga, A.; Baziard, G.; Berge, M.; Roques, C. Growth inhibition of adherent Pseudomonas aeruginosa by an N-butanoyl-L-homoserine lactone analog. Can. J. Microbiol. 2010, 56, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Allkja, J.; van Charante, F.; Aizawa, J.; Reigada, I.; Guarch-Perez, C.; Vazquez-Rodriguez, J.A.; Cos, P.; Coenye, T.; Fallarero, A.; Zaat, S.A.J.; et al. Interlaboratory study for the evaluation of three microtiter plate-based biofilm quantification methods. Sci. Rep. 2021, 11, 13779. [Google Scholar] [CrossRef] [PubMed]
- Shannon, E.; Abu-Ghannam, N. Antibacterial Derivatives of Marine Algae: An Overview of Pharmacological Mechanisms and Applications. Mar. Drugs 2016, 14, 81. [Google Scholar] [CrossRef]
- Carette, J.; Nachtergael, A.; Duez, P.; El Jaziri, M.; Rasamiravaka, T. Natural Compounds Inhibiting Pseudomonas aeruginosa Biofilm Formation by Targeting Quorum Sensing Circuitry. In Bacterial Biofilms; Dincer, S., Sümengen Özdenefe, M., Arkut, A., Eds.; IntechOpen: London, UK, 2020. [Google Scholar]
- Stiger-Pouvreau, V.; Bourgougnon, N.; Deslandes, E. Carbohydrates From Seaweeds. In Seaweed in Health and Disease Prevention; Elsevier: Amsterdam, The Netherlands, 2016; pp. 223–274. [Google Scholar]
- Paiva, L.; Lima, E.; Neto, A.I.; Marcone, M.; Baptista, J. Nutritional and Functional Bioactivity Value of Selected Azorean Macroalgae: Ulva compressa, Ulva rigida, Gelidium microdon, and Pterocladiella capillacea: Functional metabolites of selected algae. J. Food Sci. 2017, 82, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Salim, D.; Caro, P.d.; Merah, O.; Chbani, A. Control of Post-harvest Citrus Green Mold using Ulva lactuca Extracts as a Source of Active Substances. Int. J. Bio-Resour. Stress Manag. 2020, 11, 287–296. [Google Scholar] [CrossRef]
- Pantanella, F.; Valenti, P.; Natalizi, T. Analytical techniques to study microbial biofilm on abiotic surfaces: Pros and cons of the main techniques currently in use. Ann. Ig. Med. Prev. Comunita 2013, 25, 31–42. [Google Scholar] [CrossRef]
- Hidayati, J.R.; Yudiati, E.; Pringgenies, D.; Oktaviyanti, D.T.; Kusuma, A.P. Comparative Study on Antioxidant Activities, Total Phenolic Compound and Pigment Contents of Tropical Spirulina platensis, Gracilaria arcuata and Ulva lactuca Extracted in Different Solvents Polarity. E3S Web Conf. 2020, 147, 03012. [Google Scholar] [CrossRef] [Green Version]
- Dominguez, H.; Loret, E.P. Ulva lactuca, A Source of Troubles and Potential Riches. Mar. Drugs 2019, 17, 357. [Google Scholar] [CrossRef] [Green Version]
- Arsianti, A.A.; Fadilah, F.; Suid, K.; Yazid, F.; Wibisono, L.K.; Azizah, N.N.; Putrianingsih, R.; Murniasih, T.; Rasyid, A.; Pangestuti, R. Phytochemical composition and anticancer activity of seaweeds Ulva lactuca and Eucheuma cottonii against breast MCF-7 and colon HCT-116 cells. Asian J. Pharm. Clin. Res. 2016, 9, 115. [Google Scholar] [CrossRef]
- Yuvaraj, N.; Arul, V. Preliminary Screening of Anti-Biofilm, Anti-Larval Settlement and Cytotoxic Potential of Seaweeds and Seagrasses Collected from Pondicherry and Rameshwaram Coastal Line, India. WJFMS 2014, 6, 169–175. [Google Scholar] [CrossRef]
- Allkja, J.; Bjarnsholt, T.; Coenye, T.; Cos, P.; Fallarero, A.; Harrison, J.J.; Lopes, S.P.; Oliver, A.; Pereira, M.O.; Ramage, G.; et al. Minimum information guideline for spectrophotometric and fluorometric methods to assess biofilm formation in microplates. Biofilm 2020, 2, 100010. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein. Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [Green Version]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Furiga, A.; Lajoie, B.; El Hage, S.; Baziard, G.; Roques, C. Impairment of Pseudomonas aeruginosa Biofilm Resistance to Antibiotics by Combining the Drugs with a New Quorum-Sensing Inhibitor. Antimicrob. Agents Chemother. 2016, 60, 1676–1686. [Google Scholar] [CrossRef] [Green Version]
- Francolini, I.; Norris, P.; Piozzi, A.; Donelli, G.; Stoodley, P. Usnic acid, a natural antimicrobial agent able to inhibit bacterial biofilm formation on polymer surfaces. Antimicrob. Agents Chemother. 2004, 48, 4360–4365. [Google Scholar] [CrossRef] [Green Version]
- Powell, L.C.; Pritchard, M.F.; Ferguson, E.L.; Powell, K.A.; Patel, S.U.; Rye, P.D.; Sakellakou, S.-M.; Buurma, N.J.; Brilliant, C.D.; Copping, J.M.; et al. Targeted disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate oligosaccharides. NPJ Biofilms Microbiomes 2018, 4, 13. [Google Scholar] [CrossRef] [Green Version]
- Pinto, R.M.; Soares, F.A.; Reis, S.; Nunes, C.; Van Dijck, P. Innovative Strategies Toward the Disassembly of the EPS Matrix in Bacterial Biofilms. Front. Microbiol. 2020, 11, 952. [Google Scholar] [CrossRef]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents—How P. aeruginosa Can Escape Antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef] [Green Version]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, S.; Brunel, J.M.; Dubus, J.C.; Reynaud-Gaubert, M.; Rolain, J.M. Colistin: An update on the antibiotic of the 21st century. Expert Rev. Anti. Infect. Ther. 2012, 10, 917–934. [Google Scholar] [CrossRef]
- Uruen, C.; Chopo-Escuin, G.; Tommassen, J.; Mainar-Jaime, R.C.; Arenas, J. Biofilms as Promoters of Bacterial Antibiotic Resistance and Tolerance. Antibiotics 2020, 10, 3. [Google Scholar] [CrossRef]
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229. [Google Scholar] [CrossRef] [Green Version]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef]
- Zhao, F.; Wang, P.; Lucardi, R.D.; Su, Z.; Li, S. Natural Sources and Bioactivities of 2,4-Di-Tert-Butylphenol and Its Analogs. Toxins 2020, 12, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viszwapriya, D.; Prithika, U.; Deebika, S.; Balamurugan, K.; Pandian, S.K. In vitro and in vivo antibiofilm potential of 2,4-Di-tert-butylphenol from seaweed surface associated bacterium Bacillus subtilis against group A streptococcus. Microbiol. Res. 2016, 191, 19–31. [Google Scholar] [CrossRef]
- Padmavathi, A.R.; Abinaya, B.; Pandian, S.K. Phenol, 2,4-bis(1,1-dimethylethyl) of marine bacterial origin inhibits quorum sensing mediated biofilm formation in the uropathogen Serratia marcescens. Biofouling 2014, 30, 1111–1122. [Google Scholar] [CrossRef]
- Nickzad, A.; Deziel, E. The involvement of rhamnolipids in microbial cell adhesion and biofilm development—An approach for control? Lett. Appl. Microbiol. 2014, 58, 447–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohoude, M.J.; Gbaguidi, F.; Agbani, P.; Ayedoun, M.-A.; Cazaux, S.; Bouajila, J. Chemical composition and biological activities of extracts and essential oil of Boswellia dalzielii leaves. Pharm. Biol. 2017, 55, 33–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genovese, C.; D’Angeli, F.; Bellia, F.; Distefano, A.; Spampinato, M.; Attanasio, F.; Nicolosi, D.; Di Salvatore, V.; Tempera, G.; Lo Furno, D.; et al. In Vitro Antibacterial, Anti-Adhesive and Anti-Biofilm Activities of Krameria lappacea (Dombey) Burdet & B.B. Simpson Root Extract against Methicillin-Resistant Staphylococcus aureus Strains. Antibiotics 2021, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Strathmann, M.; Wingender, J.; Flemming, H.-C. Application of fluorescently labelled lectins for the visualization and biochemical characterization of polysaccharides in biofilms of Pseudomonas aeruginosa. J. Microbiol. Methods 2002, 50, 237–248. [Google Scholar] [CrossRef]





| Seaweed Species | CH P’: 0.2 | DCM P’: 3.1 | EA P’: 4.4 | MeOH P’: 5.1 | |
|---|---|---|---|---|---|
| Green alga U. lactuca | N° of repetitions | ×1 | ×2 | × 2 | ×4 |
| Color | Pale yellow | Dark green | Dark green | Dark green | |
| Yield (w/w%) | 0.2 | 0.3 | 0.1 | 12.1 | |
| Brown alga S. scoparium | N° of repetitions | ×2 | ×3 | ×3 | ×3 |
| Color | Dark yellow | Dark green | Dark green | Green | |
| Yield (w/w%) | 0.2 | 0.2 | 0.5 | 1.4 | |
| Red alga P. capillacea | N° of repetitions | ×2 | ×3 | ×3 | ×4 |
| Color | Dark yellow | Dark green | Dark green | Dark green | |
| Yield (w/w%) | 0.4 | 0.8 | 0.9 | 7.3 | |
| Seaweed Species | Nature of the Extract | CV Method | CFU Method | |
|---|---|---|---|---|
| IPCV (%) | IPCFU (%) | Log Reduction in Relation to Untreated Control | ||
| Green alga (U. lactuca) | CH | 69.4 ± 13.6 | 67.2 ± 17.2 | 0.5 ± 0.1 ** |
| DCM | 52.9 ± 9.2 | NA | 0 | |
| EA | 84.0 ± 9.6 | 44.3 ± 16.5 | 0.2 ± 0.2 NS | |
| Brown alga (S. scoparium) | DCM | 75.2 ± 15.4 | 28.1 ± 24.1 | 0.1 ± 0.1 NS |
| EA | 64.8 ± 3.6 | NA | 0 | |
| Identified Molecules | MF | MW (g/mol) | RT (min) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U. lactuca | S. scoparium | P. capillacea | ||||||||||||
| CH | DCM | EA | MeOH | CH | DCM | EA | MeOH | CH | DCM | EA | MeOH | |||
| 2,4-Dithiapentane | C3H8S2 | 108 | 7.37 | |||||||||||
| 2,4-Di-tert-butylphenol/2,5-bis(1,1-dimethylethyl)-phenol | C14H22O | 206 | 18.81 | 18.96 | 18.55 | 18.76 | 19.36 | 18.45 | 19.04 | 19.24 | 18.54 | |||
| Heptadecane | C17H36 | 240 | 20.33 | 19.72 | 20.24 | 20.04 | 19.88 | 20.5 | 20.68 | |||||
| 3,5-bis(1,1-dimethylethyl)-4-hydroxy-methyl ester benzenpropanoic acid | C18H28O3 | 292 | 23.5 | 24.5 | ||||||||||
| 2,6-bis(1,1-dimethylethyl)-4-(1-methyl-1-phenylethyl)-phenol | C23H32O | 324 | 25.66 | 26.08 | 26.2 | 26.03 | 26.59 | 25.67 | 26.29 | 26.41 | ||||
| 2,4-Bis(dimethylbenzyl)-6-t-butylphenol | C28H34O | 386 | 32.49 | 33.48 | 33.7 | 32.22 | 33.37 | 34.77 | 32.52 | 34.03 | 34.27 | |||
| 1-ethynyl-4-methyl benzene | C9H8 | 116 | 9.67 | 9.93 | ||||||||||
| 6,10,14-trimethyl-2-pentadecanone | C18H36O | 268 | 22.59 | |||||||||||
| Hexadecanoic acid methyl ester | C17H34O2 | 270 | 23.08 | 23.48 | 23.09 | 23.8 | ||||||||
| Decane | C10H22 | 142 | 7.37 | |||||||||||
| Nonanal | C9H18O | 142 | 9.39 | |||||||||||
| Isopropyl myristate | C17H34O2 | 270 | 21.83 | |||||||||||
| Tetratriacontane | C34H70 | 478 | 26.29 | |||||||||||
| Hexadecanoic acid ethyl ester | C18H36O2 | 284 | 24.61 | 24.46 | ||||||||||
| 2,4-bis(1-methyl-1-phenylethyl) phenol | C24H26O | 330 | 33.12 | 34.5 | 35.58 | 35.03 | ||||||||
| 1-ethoxy-2-propanol | C5H12O2 | 104 | 5.79 | |||||||||||
| 4-hydroxy-4-methyl-2-pentanone | C6H12O2 | 116 | 6.99 | |||||||||||
| 1-Ethoxypropane-2-yl-acetate | C7H14O3 | 146 | 7.68 | |||||||||||
| 4-(2,6,6-trimethyl-2-cyclohexen-1-yl)-3-buten-2-one | C13H20O | 192 | 18.12 | |||||||||||
| 5,6,7,7a-tetrahydro-4,4,7a-trimethyl-2(4H)-benzofuranone | C11H16O2 | 180 | 20.93 | |||||||||||
| Methyl tetradecanoate | C15H30O2 | 242 | 21.52 | |||||||||||
| 6,10,14-trimethyl-2-pentadecanone | C18H36O | 268 | 22.93 | |||||||||||
| Dibutyl phtalate | C16H22O4 | 278 | 25.28 | |||||||||||
| phytol | C20H40O | 296 | 25.81 | |||||||||||
| 3,7,11,15-tetramethylacétate-2-hexadecen-1-ol | C22H42O2 | 338 | 26.73 | |||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rima, M.; Trognon, J.; Latapie, L.; Chbani, A.; Roques, C.; El Garah, F. Seaweed Extracts: A Promising Source of Antibiofilm Agents with Distinct Mechanisms of Action against Pseudomonas aeruginosa. Mar. Drugs 2022, 20, 92. https://doi.org/10.3390/md20020092
Rima M, Trognon J, Latapie L, Chbani A, Roques C, El Garah F. Seaweed Extracts: A Promising Source of Antibiofilm Agents with Distinct Mechanisms of Action against Pseudomonas aeruginosa. Marine Drugs. 2022; 20(2):92. https://doi.org/10.3390/md20020092
Chicago/Turabian StyleRima, Maya, Jeanne Trognon, Laure Latapie, Asma Chbani, Christine Roques, and Fatima El Garah. 2022. "Seaweed Extracts: A Promising Source of Antibiofilm Agents with Distinct Mechanisms of Action against Pseudomonas aeruginosa" Marine Drugs 20, no. 2: 92. https://doi.org/10.3390/md20020092