In Silico Screening of Bioactive Compounds of Representative Seaweeds to Inhibit SARS-CoV-2 ACE2-Bound Omicron B.1.1.529 Spike Protein Trimer
Abstract
:1. Introduction
2. Materials and Methods
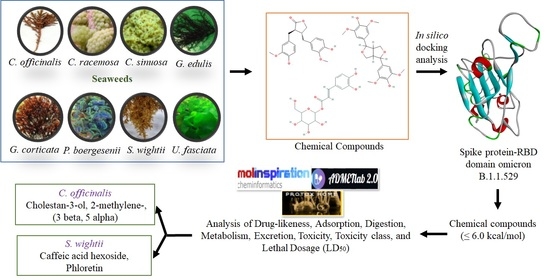
2.1. Chemical Compounds from Seaweeds
2.2. Target Preparation and Ligand Library
2.3. Molecular Docking
2.4. Evaluation of Ligands Drug-Likeness and Toxicity
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern 2021 (accessed on 19 January 2022).
- Abdullah, F.; Myers, J.; Basu, D.; Tintinger, G.; Ueckermann, V.; Mathebula, M.; Ramlall, R.; Spoor, S.; de Villiers, T.; van der Walt, Z.; et al. Decreased severity of disease during the first global omicron variant covid-19 outbreak in a large hospital in Tshwane, South Africa. Int. J. Infect. Dis. 2021, 116, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W.; et al. GISAID’s role in pandemic response. China CDC Wkly 2021, 3, 1049–1051. Available online: https://www.gisaid.org/hcov19-variants (accessed on 19 January 2022). [CrossRef] [PubMed]
- Mallapaty, S. Omicron-variant border bans ignore the evidence, say scientists. Nature 2021, 7888, 199. [Google Scholar] [CrossRef] [PubMed]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 2022, eabn7760. [Google Scholar] [CrossRef]
- Alenquer, M.; Ferreira, F.; Lousa, D.; Valério, M.; MedinaLopes, M.; Bergman, M.-L.; Goncalves, J.; Demengeot, J.; Leite, R.B.; Lilue, J.; et al. Signatures in SARS-CoV-2 spike protein conferring escape to neutralizing antibodies. PLoS Pathog. 2021, 17, e1009772. [Google Scholar] [CrossRef]
- Dupont, L.; Snell, L.B.; Graham, C.; Seow, J.; Merrick, B.; Lechmere, T.; Maguire, T.J.A.; Hallett, S.R.; Pickering, S.; Charalampous, T.; et al. Neutralizing antibody activity in convalescent sera from infection in humans with SARS-CoV-2 and variants of concern. Nat. Microbiol. 2021, 6, 1433. [Google Scholar] [CrossRef]
- Omotuyi, O.; Olubiyi, O.; Nash, O.; Afolabi, E.; Oyinloye, B.; Fatumo, S.; Femi-Oyewo, M.; Bogoro, S. SARS-CoV-2 Omicron spike glycoprotein receptor binding domain exhibits super-binder ability with ACE2 but not convalescent monoclonal antibody. Comput. Biol. Med. 2022, 142, 105226. [Google Scholar] [CrossRef]
- Kazybay, B.; Ahmad, A.; Mu, C.; Mengdesh, D.; Xie, Y. Omicron N501Y mutation among SARS-CoV-2 lineages: In silico analysis of potent binding to tyrosine kinase and hypothetical repurposed medicine. Travel Med. Infect. Dis. 2021, 45, 102242. [Google Scholar] [CrossRef]
- Ramesh, S.; Govindarajulu, M.; Parise, R.S.; Neel, L.; Shankar, T.; Patel, S.; Lowery, P.; Smith, F.; Dhanasekaran, M.; Moore, T. Emerging SARS-CoV-2 variants: A review of its mutations, its implications and vaccine efficacy. Vaccines 2021, 9, 1195. [Google Scholar] [CrossRef]
- Zhang, L.; Cui, Z.; Li, Q.; Wang, B.; Yu, Y.; Wu, J.; Nie, J.; Ding, R.; Wang, H.; Zhang, Y.; et al. Ten emerging SARS-CoV-2 spike variants exhibit variable infectivity, animal tropism, and antibody neutralization. Commun. Biol. 2021, 4, 1196. [Google Scholar] [CrossRef]
- Buss, L.F.; Prete, C., Jr.; Abrahim, C.M.M.; Mendrone, A., Jr.; Salomon, T.; Neto, C.A.; França, R.F.O.; Belotti, M.C.; Carvalho, M.P.S.S.; Costa, A.G.; et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science 2021, 371, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Ferré, V.M.; Peiffer-Smadja, N.; Visseaux, B.; Descamps, D.; Ghosn, J.; Charpentier, C. Omicron SARS-CoV-2 variant: What we know and what we don’t. Anaesth. Crit. Care Pain Med. 2021, 41, 100998. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Hong, W.; Pan, X.; Lu, G.; Wei, X. SARS-CoV-2 Omicron variant: Characteristics and prevention. MedComm 2021, 2, 838–845. [Google Scholar] [CrossRef]
- Poudel, S.; Ishak, A.; Perez-Fernandez, J.; Garcia, E.; León-Figueroa, D.A.; Romaní, L.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Highly mutated SARS-CoV-2 Omicron variant sparks significant concern among global experts—What is known so far? Travel Med. Infect. Dis. 2021, 45, 102234. [Google Scholar] [CrossRef] [PubMed]
- Aleem, A.; Samad, A.A.B.; Slenker, A.K. Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19); StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Chen, J.; Wang, R.; Gilby, N.B.; Wei, G.W. Omicron variant (B.1.1.529): Infectivity, vaccine breakthrough, and antibody resistance. J. Chem. Inf. Model 2022, 62, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Awadasseid, A.; Wu, Y.; Tanaka, Y.; Zhang, W. Effective drugs used to combat SARS-CoV-2 infection and the current status of vaccines. Biomed. Pharmacother. 2021, 137, 111330. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.F.; Shi, P.Y. Omicron: A drug developer’s perspective. Emerg. Microbes Infect. 2022, 11, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Rebours, C.; Marinho-Soriano, E.; Zertuche-González, J.A.; Hayashi, L.; Vásquez, J.A.; Kradolfer, P.; Soriano, G.; Ugarte, R.; Abreu, M.H.; Bay-Larsen, I.; et al. Seaweeds: An opportunity for wealth and sustainable livelihood for coastal communities. J. Appl. Phycol. 2014, 26, 1939–1951. [Google Scholar] [CrossRef] [Green Version]
- Vinuganesh, A.; Kumar, A.; Prakash, S.; Alotaibi, M.O.; Saleh, A.M.; Mohammed, A.E.; Beemster, G.T.S.; AbdElgawad, H. Influence of seawater acidification on biochemical composition and oxidative status of green algae Ulva Compressa. Sci. Total. Environ. 2022, 806, 150445. [Google Scholar] [CrossRef]
- Brown, E.S.; Allsopp, P.J.; Magee, P.J.; Gill, C.I.; Nitecki, S.; Strain, C.R.; McSorley, E.M. Seaweed and human health. Nutr. Rev. 2014, 72, 205–216. [Google Scholar] [CrossRef]
- Wells, M.L.; Potin, P.; Craigie, J.S.; Raven, J.A.; Merchant, S.S.; Helliwell, K.E.; Smith, A.G.; Camire, M.E.; Brawley, S.H. Algae as nutritional and functional food sources: Revisiting our understanding. J. Appl. Phycol. 2017, 29, 949–982. [Google Scholar] [CrossRef] [PubMed]
- Peñalver, R.; Lorenzo, J.M.; Ros, G.; Amarowicz, R.; Pateiro, M.; Nieto, G. Seaweeds as a functional ingredient for a healthy diet. Mar. Drugs 2020, 18, 301. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.K.; Minakuchi, M.; Wuputra, K.; Ku, C.C.; Pan, J.B.; Kuo, K.K.; Lin, Y.C.; Saito, S.; Lin, C.S.; Yokoyama, K.K. Redox control in the pathophysiology of influenza virus infection. BMC Microbiol. 2020, 20, 214. [Google Scholar] [CrossRef]
- Chernyak, B.V.; Popova, E.N.; Prikhodko, A.S.; Grebenchikov, O.A.; Zinovkina, L.A.; Zinovkin, R.A. COVID-19 and oxidative stress. Biochem. 2020, 85, 1543–1553. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, D.; Wu, J.; Chen, Y.; Wang, S. In vitro antioxidant activities of sulfated polysaccharide fractions extracted from Corallina officinalis. Int. J. Biol. Macromol. 2011, 49, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.M.; Alotaibi, B.S.; El-Sheekh, M.M. Therapeutic uses of red macroalgae. Molecules 2020, 25, 4411. [Google Scholar] [CrossRef] [PubMed]
- Yap, W.F.; Tay, V.; Tan, S.H.; Yow, Y.Y.; Chew, J. Decoding antioxidant and antibacterial potentials of Malaysian green seaweeds: Caulerpa racemosa and Caulerpa Lentillifera. Antibiotics 2019, 8, 152. [Google Scholar] [CrossRef] [Green Version]
- Pangestuti, R.; Haq, M.; Rahmadi, P.; Chun, B.S. Nutritional value and biofunctionalities of two edible green seaweeds (Ulva lactuca and Caulerpa racemosa) from Indonesia by subcritical water hydrolysis. Mar. Drugs 2021, 19, 578. [Google Scholar] [CrossRef]
- Lorente, L.; Martín, M.M.; González-Rivero, A.F.; Pérez-Cejas, A.; Argueso, M.; Perez, A.; Ramos-Gómez, L.; Solé-Violán, J.; Marcos, Y.; Ramos, J.A.; et al. Blood concentrations of proapoptotic sFas and antiapoptotic Bcl2 and COVID-19 patient mortality. Expert Rev. Mol. Diagn. 2021, 21, 837–844. [Google Scholar] [CrossRef]
- Monla, A.R.; Dassouki, Z.; Kouzayha, A.; Salma, Y.; Gali-Muhtasib, H.; Mawlawi, H. The cytotoxic and apoptotic effects of the brown algae Colpomenia sinuosa are mediated by the generation of reactive oxygen species. Molecules 2020, 25, 1993. [Google Scholar] [CrossRef]
- Rosemary, T.; Arulkumar, A.; Paramasivam, S.; Mondragon-Portocarrero, A.; Miranda, J.M. Biochemical, micronutrient and physicochemical properties of the dried red seaweeds Gracilaria edulis and Gracilaria Corticata. Molecules 2019, 24, 2225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asghar, A.; Tan, Y.C.; Shahid, M.; Yow, Y.Y.; Lahiri, C. Metabolite profiling of malaysian Gracilaria edulis reveals eplerenone as novel antibacterial compound for drug repurposing against MDR bacteria. Front. Microbiol. 2021, 12, 653562. [Google Scholar] [CrossRef]
- Ali, L.; Khan, A.L.; Al-Broumi, M.; Al-Harrasi, R.; Al-Kharusi, L.; Hussain, J.; Al-Harrasi, A. New enzyme-inhibitory triterpenoid from marine macro brown alga Padina boergesenii allender & kraft. Mar. Drugs 2017, 15, 19. [Google Scholar]
- Gora, A.H.; Sahu, N.P.; Sahoo, S.; Rehman, S.; Ahmad Dar, S.; Ahmad, I.; Agarwal, D. Effect of dietary Sargassum wightii and its fucoidan-rich extract on growth, immunity, disease resistance and antimicrobial peptide gene expression in Labeo rohita. Int. Aquat. Res. 2018, 10, 115–131. [Google Scholar] [CrossRef] [Green Version]
- Kumar, Y.; Tarafdar, A.; Kumar, D.; Badgujar, P.C. Effect of indian brown seaweed Sargassum wightii as a functional ingredient on the phytochemical content and antioxidant activity of coffee beverage. J. Food Sci. Technol. 2019, 56, 4516–4525. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar]
- Lill, M.A.; Danielson, M.L. Computer-aided drug design platform using PyMOL. J. Comput. Aided Mol. Des. 2011, 25, 13–19. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Grob, S. Slovakia. Molinspiration Cheminformatics Free Web Services. Available online: https://www.molinspiration.com (accessed on 15 January 2022).
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The single global macromolecular structure archive. Methods Mol. Biol. 2017, 1607, 627–641. [Google Scholar] [PubMed] [Green Version]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 73, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mani, J.S.; Johnson, J.B.; Steel, J.C.; Broszczak, D.A.; Neilsen, P.M.; Walsh, K.B.; Naiker, M. Natural product-derived phytochemicals as potential agents against coronaviruses: A review. Virus Res. 2020, 284, 197989. [Google Scholar] [CrossRef] [PubMed]
- Teli, D.M.; Shah, M.B.; Chhabria, M.T. In silico screening of natural compounds as potential inhibitors of SARS-CoV-2 main protease and spike RBD: Targets for COVID-19. Front. Mol. Biosci. 2021, 7, 599079. [Google Scholar] [CrossRef]
- Bharathi, M.; Sivamaruthi, B.S.; Kesika, P.; Thangaleela, S.; Chaiyasut, C. In silico screening of potential phytocompounds from several herbs against sars-cov-2 indian delta variant b.1.617.2 to inhibit the spike glycoprotein trimer. Appl. Sci. 2022, 12, 665. [Google Scholar] [CrossRef]
- Jayawardena, R.; Sooriyaarachchi, P.; Chourdakis, M.; Jeewandara, C.; Ranasinghe, P. Enhancing immunity in viral infections, with special emphasis on COVID-19: A review. Diabetes Metab. Syndr. 2020, 14, 367–382. [Google Scholar] [CrossRef]
- Leandro, A.; Pacheco, D.; Cotas, J.; Marques, J.C.; Pereira, L.; Gonçalves, A.M.M. Seaweed’s bioactive candidate compounds to food industry and global food security. Life 2020, 10, 140. [Google Scholar] [CrossRef]
- Ahmed, M.H.; Hassan, A.; Molnár, J. The role of micronutrients to support immunity for covid-19 prevention. Rev. Bras. Farmacogn. 2021, 31, 361–374. [Google Scholar] [CrossRef]
- Tamama, K. Potential benefits of dietary seaweeds as protection against COVID-19. Nutr. Rev. 2021, 79, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.G.; Tuvikene, R. Potential antiviral properties of industrially important marine algal polysaccharides and their significance in fighting a future viral pandemic. Viruses 2021, 13, 1817. [Google Scholar]
- McMahan, K.; Yu, J.; Mercado, N.B.; Loos, C.; Tostanoski, L.H.; Chandrashekar, A.; Liu, J.; Peter, L.; Atyeo, C.; Zhu, A.; et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2021, 590, 630–634. [Google Scholar] [CrossRef]
- Collie, S.; Champion, J.; Moultrie, H.; Bekker, L.G.; Gray, G. Effectiveness of BNT162b2 vaccine against omicron variant in South Africa. N. Engl. J. Med. 2022, 386, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Peiris, M.; Cheng, S.; Mok, C.K.P.; Leung, Y.; Ng, S.; Chan, K.; Ko, F.; Yiu, K.; Lam, B.; Lau, E.; et al. Neutralizing antibody titres to SARS-CoV-2 Omicron variant and wild-type virus in those with past infection or vaccinated or boosted with mRNA BNT162b2 or inactivated CoronaVac vaccines. Res. Sq. 2022, rs.3.rs-1207071, Preprint. [Google Scholar] [CrossRef]
- Durojaiye, A.B.; Clarke, J.D.; Stamatiades, G.A.; Wang, C. Repurposing cefuroxime for treatment of COVID-19: A scoping review of in silico studies. J. Biomol. Struct. Dyn. 2021, 39, 4547–4554. [Google Scholar] [CrossRef]
- Adebisi, Y.A.; Jimoh, N.D.; Ogunkola, I.O.; Uwizeyimana, T.; Olayemi, A.H.; Ukor, N.A.; Lucero-Prisno, D.E., 3rd. The use of antibiotics in COVID-19 management: A rapid review of national treatment guidelines in 10 African countries. Trop. Med. Health. 2021, 49, 51. [Google Scholar] [CrossRef]
- Mustafa, L.; Tolaj, I.; Baftiu, N.; Fejza, H. Use of antibiotics in COVID-19 ICU patients. J. Infect. Dev. Ctries. 2021, 15, 501–505. [Google Scholar] [CrossRef]
- Alberca, R.W.; Teixeira, F.M.E.; Beserra, D.R.; de Oliveira, E.A.; Andrade, M.M.S.; Pietrobon, A.J.; Sato, M.N. Perspective: The potential effects of naringenin in COVID-19. Front. Immunol. 2020, 11, 570919. [Google Scholar] [CrossRef]
- Shawan, M.M.A.K.; Halder, S.K.; Hasan, M.A. Luteolin and abyssinone II as potential inhibitors of SARS-CoV-2: An in silico molecular modeling approach in battling the COVID-19 outbreak. Bull. Natl. Res. Cent. 2021, 45, 27. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, L.; Mo, M.; Liu, T.; Wu, C.; Gong, C.; Lu, K.; Gong, L.; Zhu, W.; Xu, Z. The effect of the multiple mutations in Omicron RBD on its binding to human ACE2 receptor and immune evasion: An investigation of molecular dynamics simulations. ChemRxiv. Camb. Camb. Open Engag. 2021. Preprint. [Google Scholar] [CrossRef]
- Kulkarni, S.A.; Krishnan, S.B.B.; Chandrasekhar, B.; Banerjee, K.; Sohn, H.; Madhavan, T. Characterization of phytochemicals in Ulva intestinalis L. and their action against SARS-CoV-2 spike glycoprotein receptor-binding domain. Front. Chem. 2021, 27, 735768. [Google Scholar] [CrossRef] [PubMed]
- Adem, Ş.; Eyupoglu, V.; Sarfraz, I.; Rasul, A.; Zahoor, A.F.; Ali, M.; Abdalla, M.; Ibrahim, I.M.; Elfiky, A.A. Caffeic acid derivatives (CAFDs) as inhibitors of SARS-CoV-2: CAFDs-based functional foods as a potential alternative approach to combat COVID-19. Phytomedicine 2021, 85, 153310. [Google Scholar] [CrossRef] [PubMed]
- Utsunomiya, H.; Ichinose, M.; Ikeda, K.; Uozaki, M.; Morishita, J.; Kuwahara, T.; Koyama, A.H.; Yamasaki, H. Inhibition by caffeic acid of the influenza A virus multiplication in vitro. Int. J. Mol. Med. 2014, 34, 1020–1024. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, M.; Shirasago, Y.; Ando, S.; Shimojima, M.; Saijo, M.; Fukasawa, M. Caffeic acid, a coffee-related organic acid, inhibits infection by severe fever with thrombocytopenia syndrome virus in vitro. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2018, 24, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Langland, J.; Jacobs, B.; Wagner, C.E.; Ruiz, G.; Cahill, T.M. Antiviral activity of metal chelates of caffeic acid and similar compounds towards herpes simplex, VSV-Ebola pseudotyped and vaccinia viruses. Antivir. Res. 2018, 160, 143–150. [Google Scholar] [CrossRef]
- Muhammad, Y.; Kani, Y.A.; Iliya, S.; Muhammad, J.B.; Binji, A.; Ahmad, E.A.; Kabir, M.B.; Bindawa, U.K.; Ahmed, A. Deficiency of antioxidants and increased oxidative stress in COVID-19 patients: A cross-sectional comparative study in Jigawa, Northwestern Nigeria. SAGE Open Med. 2021, 9, 2050312121991246. [Google Scholar] [CrossRef] [PubMed]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef] [Green Version]
- Beltrán-García, J.; Osca-Verdegal, R.; Pallardó, F.V.; Ferreres, J.; Rodríguez, M.; Mulet, S.; Sanchis-Gomar, F.; Carbonell, N.; García-Giménez, J.L. Oxidative stress and inflammation in COVID-19-associated sepsis: The potential role of anti-oxidant therapy in avoiding disease progression. Antioxidants 2020, 9, 936. [Google Scholar] [CrossRef]
- Behzad., S.; Sureda, A.; Barreca, D.; Nabavi, S.F.; Rastrelli, L.; Nabavi, S.M. Health effects of phloretin: From chemistry to medicine. Phytochem. Rev. 2017, 16, 527–533. [Google Scholar] [CrossRef]
- Fadaka, A.O.; Sibuyi, N.R.S.; Martin, D.R.; Klein, A.; Madiehe, A.; Meyer, M. Development of effective therapeutic molecule from natural sources against coronavirus protease. Int. J. Mol. Sci. 2021, 22, 9431. [Google Scholar] [CrossRef]
- Guan, L.; Yang, H.; Cai, Y.; Sun, L.; Di, P.; Li, W.; Liu, G.; Tang, Y. ADMET-score—A comprehensive scoring function for evaluation of chemical drug-likeness. Medchemcomm 2018, 10, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Benkerroum, N. Chronic and acute toxicities of aflatoxins: Mechanisms of action. Int. J. Environ. Res. Public Health. 2020, 17, 423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Dong, G.; Sheng, C. Structural simplification: An efficient strategy in lead optimization. Acta. Pharm. Sin. B. 2019, 9, 880–901. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- Jayaraj, V.; Suhanya, R.; Vijayasarathy, M.; Anandagopu, P.; Rajasekaran, E. Role of large hydrophobic residues in proteins. Bioinformation 2009, 3, 409–412. [Google Scholar] [CrossRef]
- Weinstein, J.Y.; Elazar, A.; Fleishman, S.J. A lipophilicity-based energy function for membrane-protein modelling and design. PLoS Comput. Biol. 2019, 15, e1007318. [Google Scholar] [CrossRef] [Green Version]
- Gowder, M.S.; Chatterjee, J.; Chaudhuri, T.; Paul, K. Prediction and analysis of surface hydrophobic residues in tertiary structure of proteins. Sci. World J. 2014, 2014, 971258. [Google Scholar]
- Benet, L.Z.; Hosey, C.M.; Ursu, O.; Oprea, T.I. BDDCS, the Rule of 5 and drugability. Adv. Drug. Deliv. Rev. 2016, 101, 89–98. [Google Scholar] [CrossRef] [Green Version]




| Seaweeds Name | Compound Name | Molecular Formula | Mol. Weight (g/mol) | PubChem ID |
|---|---|---|---|---|
| Standard drug | Ceftriaxone | C18H18N8O7S3 | 554.6 | 5479530 |
| Cefuroxime | C16H16N4O8S | 424.4 | 5479529 | |
| C. officinalis | 3’,8,8’-Trimethoxy-3-piperidin-1-yl2,2’-binaphthyl-1,1’,4,4’-tetrone | C28H25NO7 | 487.5 | 590815 |
| Cholestan-3-ol, 2-methylene-, (3beta, 5 alpha) | C28H48O | 400.7 | 22213932 | |
| C.racemosa | Glucobrassicin | C16H20N2O9S2 | 448.5 | 656506 |
| C. sativa L | Matairesinol | C20H22O6 | 358.4 | 119205 |
| Naringenin | C15H12O5 | 272.25 | 932 | |
| Syringaresinol | C22H26O8 | 418.4 | 100067 | |
| G. corticate | Bicyclo[3.2.1]oct-3-en-2-one, 3,8-dihydroxy-7-(7-methoxy-1,3-benzodioxol-5-yl)-6-methyl-5-(2-propenyl)-, [1S-(6-endo,7-exo,8-syn)]- | C20H22O6 | 358.39 | 101282028 |
| Cholesta-8,24-dien-3-ol, 4-methyl-, (3.beta.,4.alpha.)- | C28H46O | 398.7 | 22212496 | |
| P.boergesenii | Glucobrassicin | C16H20N2O9S2 | 448.5 | 656506 |
| Glycitein | C16H12O5 | 284.26 | 5317750 | |
| Matairesinol | C20H22O6 | 358.4 | 119205 | |
| Naringenin | C15H12O5 | 272.25 | 932 | |
| Pyrano [4,3-b] benzopyran-1,9-dione, 5amethoxy-9amethyl-3-(1-propenyl) perhydro | C17H24O5 | 308.4 | 5364482 | |
| Syringaresinol | C22H26O8 | 418.4 | 100067 | |
| S. wightii | 5-p-coumaroylquinic acid | C16H18O8 | 338.31 | 6441280 |
| Caffeic acid hexoside | C15H18O9 | 342.3 | 6124135 | |
| Phloretin | C15H14O5 | 274.27 | 4788 | |
| Quercetin-3-O-arabinoglucoside | C26H28O16 | 596.5 | 5484066 |
| Seaweeds | Chemical Compound | Binding Affinity | RMSD (Å) | H/C-H Bond Interaction | Interaction Distances | Hydrophobic Interaction | Alkyl Interaction | Pi-Sigma /Cation Stacked Interaction |
|---|---|---|---|---|---|---|---|---|
| Standard drug | Ceftriaxone | −7.1 | 43.189 | ARG403, ASN417, TYR453, SER494, SER496, TYR501 | 5.36, 4.21, 6.03, 3.24, 3.59, 6.23 | ASP405, GLU406, ARG408, GLN409, LEU455, ARG493 | - | ARG403, HIS505 |
| Cefuroxime | −5.3 | 2.483 | THR376, GLY404 *, ARG408, TYR508 | 4.52, 5.06, 4.71, 5.71 | PHE375, ASN437 | ARG408, VAL503 | - | |
| C. officinalis | 3’,8,8’-Trimethoxy-3-piperidin-1-yl2,2’-binaphthyl-1,1’,4,4’-tetrone | −6.9 | 1.897 | - | - | ARG355, TYR396, ASP428, PHE429, THR430, SER514, PHE515, LEU517, LEU518 | PRO426, PRO463 | PHE464, GLU516 |
| Cholestan-3-ol, 2-methylene-, (3beta, 5 alpha) | −6.0 | 3.074 | SER494 | 4.22 | GLY482, THR470 | LEU452, TYR449, ILE472, ALA484, PHE490 | PHE490 | |
| C. racemosa | Glucobrassicin | −6.8 | 1.521 | ARG457, ARG466, ASP467, ASP467 * | 4.45, 6.50, 3.18, 5.49 | ARG454, PHE456, SER459, GLU465, ILE468, SER469, TYR473, PRO491 | ARG457, LYS458 | ARG457, ASP467, GLU471 |
| G. corticata | Bicyclo[3.2.1]oct-3-en-2-one, 3,8-dihydroxy-1-methoxy-7-(7-methoxy-1,3-benzodioxol-5-yl)-6-methyl-5 | −6.3 | 20.322 | THR430 | 4.16 | ARG355, ASP428, SER514, PHE515, GLU516, LEU517 | TYR396, PRO426, PHE429, PRO463, PHE464 | - |
| Cholesta-8,24-dien-3-ol, 4-methyl-, (3.beta.,4.alpha.)- | −6.8 | 1.544 | - | - | TYR396, ASP428, THR430, GLU465, SER514, PHE515, GLU516 | PRO426, LYS462, PRO463, PHE464 | - | |
| P. boergesenii | Glucobrassicin | −6.8 | 1.521 | ARG454, LYS458, SER459, SER469 | 5.00, 4.44, 2.38, 3.25 | PHE456, ARG457, TYR473, PRO491 | - | ASP467, GLU471 |
| Glycitein | −6.1 | 11.668 | THR376, ASP405 * | 4.71, 4.5 | GLY404, ARG408, VAL503, GLY504 | VAL407 | TYR508, PHE375 | |
| Matairesinol | −6.0 | 1.707 | ARG355, PHE464 *, SER514 | 6.65, 4.98, 3.72 | TYR396, ASP428, PHE429, THR430, PRO463, PHE515, GLU516, LEU517 | PRO426 | - | |
| Naringenin | −6.4 | 20.209 | ASN437, LYS440, LEU441 | 4.09, 4.44, 4.14 | ASN343, PHE374, PHE375, SER438, ASN439 | PRO373, LYS440 | TRP436 | |
| Pyrano [4,3-b] benzopyran-1,9-dione, 5a-methoxy-9a-methyl-3-(1-propenyl) perhydro | −6.1 | 29.617 | SER496, TYR501 | 3.39, 5.24 | ARG403, TYR453, TYR495, GLY502 | HIS505 | TYR501, HIS505 | |
| Syringaresinol | −6.4 | 0.44 | ARG355, ASP427 *, PRO463 *, PHE515 *, GLU516 | 6.88, 4.19, 4.81, 7.35, 3.78 | ASP428, THR430, SER514 | TYR396, PRO426, LYS462, PRO463 | PHE464 | |
| S. wightii | 5-p-coumaroylquinic acid | −6.0 | 4.96 | ARG403, ASN417, TYR453, SER496 | 5.43, [2.96, 4.87], 5.62, 3.42 | GLN409, GLY416, ILE418, LEU455, SER494, TYR495, TYR501, HIS505 | - | - |
| Caffeic acid hexoside | −6.4 | 2.82 | ARG403, GLU406, ASN417, TYR453, SER496 | [5.38, 6.24], 3.95, 4.96, 5.76, [3.34, 3.36] | ASP405, ARG408, GLN409, ILE418, LEU455, TYR495, SER494, TYR501, HIS505 | - | - | |
| Phloretin | −6.3 | 0.061 | TYR501, SER496, TYR453 | 5.37, 1.49, 5.90 | ARG403, TYR495, PHE497, THR500, GLY502 | - | TYR501, HIS505 | |
| Quercetin-3-O-arabinoglucoside | −6.1 | 2.248 | ASN331, THR333, GLY526, PRO527 *, LYS528C | 5.09, [3.47, 3.38], [3.93, 4.29], 4.60, 5.10 | PRO330, ILE332, CYS361, THR523 | VAL362, CYS525 | ASN360 |
| Seaweeds | Chemical Compounds | Drug−Likeness | Toxicity Analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mi LogP | TPSA | natoms | nON | nOHNH | #Violations | Intestinal Absorption | Hepato Toxicity | Carcino Genicity | Immuno toxicity | Muta Genicity | Cyto toxicity | LD50 (mg/kg) | TC | ||
| Standard Drug | Cefuroxime | −0.98 | 173.77 | 29 | 12 | 4 | 1 | 0.065 | 0.66(Mild) | 0.50 (Mod) | 0.99(−) | 0.76(−) | 0.54(Mod) | 10,000 | VI |
| C. officinalis | 3’,8,8’−Trimethoxy−3−piperidin−1−yl2,2’−binaphthyl−1,1’,4,4’−tetrone | 3.99 | 99.22 | 36 | 8 | 0 | 0 | 0.606 | 0.83(−) | 0.55(Mod) | 0.70(+) | 0.56(Mod) | 0.58(Mod) | 400 | IV |
| Cholestan−3−ol, 2−methylene−, (3beta, 5 alpha) | 8.11 | 20.23 | 29 | 1 | 1 | 1 | 0.922 | 0.94(−) | 0.62(Mild) | 0.98(+) | 0.94(−) | 0.94(−) | 5000 | V | |
| G. corticate P.boergesenii | Cholesta−8,24−dien−3−ol, 4−methyl−, (3.beta.,4.alpha.)− | 7.96 | 20.23 | 29 | 1 | 1 | 1 | 0.931 | 0.82(−) | 0.58(Mod) | 0.97(+) | 0.94(−) | 0.96(−) | 2000 | IV |
| Syringaresinol | 2.62 | 95.86 | 30 | 8 | 2 | 0 | 0.599 | 0.87(−) | 0.54(Mod) | 0.95(+) | 0.84(−) | 0.99(−) | 1500 | IV | |
| S. wightii | Caffeic acid hexoside | −0.77 | 156.91 | 24 | 9 | 6 | 1 | 0.221 | 0.82(−) | 0.76(−) | 0.95(+) | 0.78(−) | 0.87(−) | 5000 | V |
| Phloretin | 2.66 | 97.98 | 20 | 5 | 4 | 0 | 0.427 | 0.63(Mod) | 0.72(mild) | 0.98(−) | 0.88(−) | 0.82(−) | 500 | IV | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bharathi, M.; Sivamaruthi, B.S.; Kesika, P.; Thangaleela, S.; Chaiyasut, C. In Silico Screening of Bioactive Compounds of Representative Seaweeds to Inhibit SARS-CoV-2 ACE2-Bound Omicron B.1.1.529 Spike Protein Trimer. Mar. Drugs 2022, 20, 148. https://doi.org/10.3390/md20020148
Bharathi M, Sivamaruthi BS, Kesika P, Thangaleela S, Chaiyasut C. In Silico Screening of Bioactive Compounds of Representative Seaweeds to Inhibit SARS-CoV-2 ACE2-Bound Omicron B.1.1.529 Spike Protein Trimer. Marine Drugs. 2022; 20(2):148. https://doi.org/10.3390/md20020148
Chicago/Turabian StyleBharathi, Muruganantham, Bhagavathi Sundaram Sivamaruthi, Periyanaina Kesika, Subramanian Thangaleela, and Chaiyavat Chaiyasut. 2022. "In Silico Screening of Bioactive Compounds of Representative Seaweeds to Inhibit SARS-CoV-2 ACE2-Bound Omicron B.1.1.529 Spike Protein Trimer" Marine Drugs 20, no. 2: 148. https://doi.org/10.3390/md20020148