Isolation and Structural Elucidation of New Amphidinol Analogues from Amphidinium carterae Cultivated in a Pilot-Scale Photobioreactor
Abstract
:1. Introduction
2. Results and Discussion
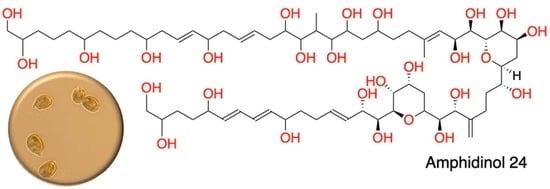
2.1. Isolation and Structural Elucidation of AMs
2.2. Hemolytic Activity
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Biological Material
3.3. Cultivation in the LED-Based Bubble Column PBR
3.4. Extraction and Chromatographic Separation
3.5. Hemolytic Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References and Note
- Gallardo-Rodríguez, J.J.; Sánchez-Mirón, A.; García-Camacho, F.; López-rosales, L.; Chisti, Y.; Molina-Grima, E. Bioactives from microalgal dinoflagellates. Biotechnol. Adv. 2012, 30, 1673–1684. [Google Scholar] [CrossRef]
- Assunção, J.; Guedes, A.C.; Malcata, F.X. Biotechnological and pharmacological applications of biotoxins and other bioactive molecules from dinoflagellates. Mar. Drugs 2017, 15, 393. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, Y. Microalgae as a Drug Source. In Drugs from the Sea; Fusetani, N., Ed.; Karger Medical and Scientific Publishers: Basel, Switzerland, 2000; pp. 30–45. [Google Scholar]
- Gallardo-Rodríguez, J.J.; López-rosales, L.; Sánchez-Mirón, A.; García-Camacho, F.; Molina-Grima, E.; Chalmers, J.J. New insights into shear-sensitivity in dinoflagellate microalgae. Bioresour. Technol. 2016, 200, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Rodríguez, J.J.; Sánchez-Mirón, A.; García-Camacho, F.; López-rosales, L.; Chisti, Y.; Molina-Grima, E. Causes of shear sensitivity of the toxic dinoflagellate Protoceratium reticulatum. Biotechnol. Prog. 2009, 25, 792–800. [Google Scholar] [CrossRef]
- Molina-Miras, A.; Morales-Amador, A.; de Vera, C.R.; López-Rosales, L.; Sánchez-Mirón, A.; Souto, M.L.; Fernández, J.J.; Norte, M.; García-Camacho, F.; Molina-Grima, E. A pilot-scale bioprocess to produce amphidinols from the marine microalga Amphidinium carterae: Isolation of a novel analogue. Algal Res. 2018, 31, 87–98. [Google Scholar] [CrossRef]
- Molina-Miras, A.; López-Rosales, L.; Sánchez-Mirón, A.; Cerón-García, M.C.; Seoane-Parra, S.; García-Camacho, F.; Molina-Grima, E. Long-term culture of the marine dinoflagellate microalga Amphidinium carterae in an indoor LED-lighted raceway photobioreactor: Production of carotenoids and fatty acids. Bioresour. Technol. 2018, 265, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Abreu, A.C.; Molina-Miras, A.; Aguilera-Saez, L.M.; López-Rosales, L.; Cerón-García, M.C.; Sánchez-Mirón, A.; Olmo-García, L.; Carrasco-Panorbo, A.; García-Camacho, F.; Molina-Grima, E.; et al. Production of amphidinols and other bioproducts of interest by the marine microalga Amphidinium carterae unraveled by nuclear magnetic resonance metabolomics approach coupled to multivariate data analysis. J. Agric. Food Chem. 2019, 67, 9667–9682. [Google Scholar] [CrossRef] [PubMed]
- Molina-Miras, A.; López-Rosales, L.; Cerón-García, M.C.; Sánchez-Mirón, A.; Olivera-Gálvez, A.; García-Camacho, F.; Molina-Grima, E. Acclimation of the microalga Amphidinium carterae to different nitrogen sources: Potential application in the treatment of marine aquaculture effluents. J. Appl. Phycol. 2020, 32, 1075–1094. [Google Scholar] [CrossRef]
- Kobayashi, J.; Kubota, T. Bioactive metabolites from marine dinoflagellates. In Comprehensive Natural Products II Chemistry and Biology; Elsevier: Oxford, UK, 2010; Volume 2, pp. 263–325. [Google Scholar] [CrossRef]
- Kobayashi, J.; Kubota, T. Bioactive macrolides and polyketides from marine dinoflagellates of the genus Amphidinium. J. Nat. Prod. 2007, 70, 451–460. [Google Scholar] [CrossRef]
- Li, W.-S.; Yan, R.-J.; Yu, Y.; Shi, Z.; Mándi, A.; Shen, L.; Kurtán, T.; Wu, J. Determination of the absolute configuration of Super-Carbon-Chain compounds by a combined chemical, spectroscopic, and computational approach: Gibbosols A and B. Angew. Chem. 2020, 59, 13028–13036. [Google Scholar] [CrossRef]
- Martínez, K.A.; Lauritano, C.; Druka, D.; Romano, G.; Grohmann, T.; Jaspars, M.; Martín, J.; Díaz, C.; Cautain, B.; de la Cruz, M.; et al. Amphidinol 22, a new cytotoxic and antifungal amphidinol from the dinoflagellate Amphidinium carterae. Mar. Drugs 2019, 17, 385. [Google Scholar] [CrossRef] [Green Version]
- Wellkamp, M.; García-Camacho, F.; Durán-Riveroll, L.M.; Tebben, J.; Tillmann, U.; Krock, B. LC-MS/MS method development for the discovery and identification of amphidinols produced by Amphidinium. Mar. Drugs 2020, 18, 497. [Google Scholar] [CrossRef]
- Minamida, M.; Kumagai, K.; Ulanova, D.; Akakabe, M.; Konishi, Y.; Tominaga, A.; Tanaka, H.; Tsuda, M.; Fukishi, E.; Kawabata, J.; et al. Amphirionin-4 with potent proliferation-promoting activity on bone marrow stromal cells from a marine dinoflagellate Amphidinium species. Org. Lett. 2014, 16, 4858–4861. [Google Scholar] [CrossRef] [PubMed]
- Cutignano, A.; Nuzzo, G.; Sardo, A.; Fontana, A. The missing piece in biosynthesis of amphidinols: First evidence of glycolate as a starter unit in new polyketides from Amphidinium carterae. Mar. Drugs 2017, 15, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J. Method for Culturing Mycoplasma Contamination-Free Cells and Method for Removing Mycoplasma Contamination of Cells. U.S. Patent 8,815,565, 26 August 2014. [Google Scholar]
- Espiritu, R.A. Membrane permeabilizing action of amphidinol 3 and theonellamide A in raft-forming lipid mixtures. Z. Naturforsch. C J. Biosci. 2017, 72, 43–48. [Google Scholar] [CrossRef] [PubMed]
- We have renamed our metabolite amphidinol 20 in reference 6 as amphidinol 20B due to its publication coincided in time with that of another derivative called with the same name but which came out in press before ours (Satake, M.; Cornelio, K.; Hanashima, S.; Malabed, R.; Murata, M.; Matsumori, N.; Zhang, H.; Hayashi, F.; Mori, S.; Kim, J.S.; et al. Structures of the largest amphidinol homologues from the dinoflagellate Amphidinium carterae and structure-activity relationships. J. Nat. Prod. 2017, 80, 2883–2888. [Google Scholar] [CrossRef])
- Kubota, T.; Takahashi, A.; Tsuda, M.; Kobayashi, J. Luteophanol D, new polyhydroxyl metabolite from marine dinoflagellate Amphidinium sp. Mar. Drugs 2005, 3, 113–118. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.-C.; Zhao, D.; Guo, Y.-W.; Wu, H.-M.; Lin, L.-P.; Wang, Z.-H.; Ding, J.; Lin, Y.-S. Lingshuiol, a novel polyhydroxyl compound with strongly cytotoxic activity from the marine dinoflagellate Amphidinium sp. Bioorg. Med. Chem. Lett. 2004, 14, 3117–3120. [Google Scholar] [CrossRef]
- Wakamiya, Y.; Ebine, M.; Murayama, M.; Omizu, H.; Matsumori, N.; Murata, M.; Oishi, T. Synthesis and stereochemical revision of the C31-C67 section of amphidinol 3. Angew. Chem. 2018, 130, 6168–6172. [Google Scholar] [CrossRef]
- Molina-Miras, A.; López-Rosales, L.; Cerón-García, M.; Sánchez-Mirón, A.; García-Camacho, F.; Contreras-Gómez, A.; Molina-Grima, E. A new approach to finding optimal centrifugation conditions for shear-sensitive microalgae. Algal Res. 2019, 44, 101677. [Google Scholar] [CrossRef]
- Leung, W.W.-F. Centrifugal Separations in Biotechnology; Butterworth-Heinemann: Oxford, UK, 2020. [Google Scholar]
- Juhl, A.R.; Trainer, V.L.; Latz, M.I. Effect of fluid shear and irradiance on population growth and cellular toxin content of the dinoflagellate Alexandrium fundyense. Limnol. Oceanogr. 2001, 46, 758–764. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, J.G.; Mirón, A.S.; Camacho, F.G.; García, M.C.; Belarbi, E.; Chisti, Y.; Grima, E.M. Carboxymethyl cellulose and Pluronic F68 protect the dinoflagellate Protoceratium reticulatum against shear-associated damage. Bioprocess Biosyst. Eng. 2011, 34, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Nagai, S.; Suzuki, T.; Nishikawa, T.; Kamiyama, T. Differences in the production and excretion kinetics of okadaic acid, dinophysistoxin-1, and pectenotoxin-2 between cultures of Dinophysis acuminata and Dinophysis fortii isolated from western Japan. J. Phycol. 2011, 47, 1326–1337. [Google Scholar] [CrossRef]
- Nielsen, L.T.; Krock, B.; Hansen, P.J. Production and excretion of okadaic acid, pectenotoxin-2 and a novel dinophysistoxin from the DSP-causing marine dinoflagellate Dinophysis acuta—Effects of light, food availability and growth phase. Harmful Algae 2013, 23, 34–45. [Google Scholar] [CrossRef]
- Napolitano, J.G.; Norte, M.; Padrón, J.M.; Fernández, J.J.; Hernández Daranas, A. Belizeanolide, a cytotoxic macrolide from the dinoflagellate Prorocentrum belizeanum. Angew. Chem. Int. Ed. 2009, 48, 796–799. [Google Scholar] [CrossRef]
- Rodríguez, F.; Riobó, P.; Crespín, G.D.; Daranas, A.H.; de Vera, C.R.; Norte, M.; Fernández, J.J.; Fraga, S. The toxic benthic dinoflagellate Prorocentrum maculosum Faust is a synonym of Prorocentrum hoffmannianum Faust. Harmful Algae 2018, 78, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Morsy, N.; Houdai, T.; Konoki, K.; Matsumori, N.; Oishi, T.; Murata, M. Effects of lipid constituents on membrane-permeabilizing activity of amphidinols. Bioorg. Med. Chem. 2008, 16, 3084–3090. [Google Scholar] [CrossRef]
- Swasono, R.T.; Mouri, R.; Morsy, R.; Matsumoto, N.; Oishi, T.; Murata, M. Sterol effect on interaction between amphidinol 3 and liposomal membrane as evidenced by surface plasmon resonance. Bioorg. Med. Chem. Lett. 2010, 20, 2215–2218. [Google Scholar] [CrossRef]
- Iwamoto, M.; Sumino, A.; Shimada, E.; Kinoshita, M.; Matsumori, N.; Oiki, S. Channel formation and membrane deformation via sterol-aided polymorphism of amphidinol 3. Sci. Rep. 2017, 7, 10782. [Google Scholar] [CrossRef]
- Houdai, T.; Matsuoka, M.; Matsumori, N.; Murata, M. Membrane-permeabilizing activities of amphidinol 3, polyene-polyhydroxy antifungal from a marine dinoflagellate. Biochim. Biophys. Acta 2004, 1667, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Espiritu, R.A.; Matsumori, N.; Tsuda, M.; Murata, M. Direct and stereospecific interaction of amphidinol 3 with sterol in lipid bilayers. Biochemistry 2014, 53, 3287–3293. [Google Scholar] [CrossRef]
- Houdai, T.; Matsumori, N.; Murata, M. Structure of membrane-bound amphidinol 3 in isotropic small bicelles. Org. Lett. 2008, 10, 4191–4194. [Google Scholar] [CrossRef]
- Houdai, T.; Matsuoka, S.; Morsy, N.; Matsumori, N.; Satake, M.; Murata, M. Hairpin conformation of amphidinols possibly accounting for potent membrane permeabilizing activities. Tetrahedron 2005, 61, 2795–2802. [Google Scholar] [CrossRef]
- Swasono, R.T.; Kanemoto, M.; Matsumori, N.; Oishi, T.; Murata, M. Structural reevaluations of amphidinol 3, a potent antifungal compound from dinoflagellate. Heterocycles 2011, 82, 1359–1369. [Google Scholar] [CrossRef]
- Murata, M.; Matsumoro, N.; Konoki, K.; Oishi, T. Structural features of dinoflagellate toxins underlying biological activity as viewed by NMR. Bull. Chem. Soc. Jpn. 2008, 3, 307–319. [Google Scholar] [CrossRef] [Green Version]
- Paul, G.K.; Matsumori, N.; Konoki, K.; Murata, M.; Tachibana, K. Chemical structures of amphidinols 5 and 6 isolated from marine dinoflagellate Amphidinium klebsii and their cholesterol-dependent membrane disruption. J. Mar. Biotechnol. 1997, 5, 124–128. [Google Scholar]
- Eschbach, E.; Scharsack, J.P.; John, U.; Medlin, L.K. Improved erythrocyte lysis assay in microtitre plates for sensitive detection and efficient measurement of haemolytic compounds from ichthyotoxic algae. J. Appl. Toxicol. 2001, 21, 513–519. [Google Scholar] [CrossRef]
- Morsy, N.; Konoki, K.; Houdai, T.; Matsumori, N.; Oishi, T.; Murata, M.; Aimoto, S. Roles of integral protein in membrane permeabilization by amphidinols. Biochim. Biophys. Acta 2008, 1778, 1453–1459. [Google Scholar] [CrossRef] [Green Version]
- Morsy, N.; Houdai, T.; Matsuoka, S.; Matsumori, N.; Adachi, S.; Oishi, T.; Murata, M.; Iwashita, T.; Fujita, T. Structures of new amphidinols with truncated polyhydroxyl chain and their membrane-permeabilizing activities. Bioorg. Med. Chem. 2006, 14, 6548–6554. [Google Scholar] [CrossRef]
- Wakamiya, Y.; Ebine, M.; Matsumori, N.; Oishi, T. Total synthesis of amphidinol 3: A general strategy for synthesizing amphidinol analogues and structure−activity relationship study. J. Am. Chem. Soc. 2020, 142, 3472–3478. [Google Scholar] [CrossRef] [PubMed]
- Matsumori, N.; Murata, M. 3D structures of membrane-associated small molecules as determined by isotropic bicelles. Nat. Prod. Rep. 2010, 27, 1480–1492. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Ishibashi, M.; Nakamichi, H.; Kosaka, T. Ishikawa, T.; Kobayashi, J. Luteophanol A, a new polyhydroxyl compound from symbiotic marine dinoflagellate Amphidinium sp. J. Org. Chem. 1997, 62, 3820–3823. [Google Scholar] [CrossRef]
- Echigoya, R.; Rhodes, L.; Oshima, Y.; Satake, M. The structures of five new antifungal and hemolytic amphidinol analogs from Amphidinium carterae collected in New Zealand. Harmful Algae 2005, 4, 383–389. [Google Scholar] [CrossRef]
- Keller, M.D.; Selvin, R.C.; Claus, W.; Guillard, R.R.L. Media for the culture of oceanic ultraplankton. J. Phycol. 1987, 23, 633–638. [Google Scholar] [CrossRef]
- López-Rosales, L.; García-Camacho, F.; Sánchez-Mirón, A.; Beato, E.M.; Chisti, Y.; Grima, E.M. Pilot-scale bubble column photobioreactor culture of a marine dinoflagellate microalga illuminated with light emission diodes. Bioresour. Technol. 2016, 216, 845–855. [Google Scholar] [CrossRef]







| AM24 (1) | AM25 (2) | AM26 (3) | ||||
|---|---|---|---|---|---|---|
| Carbon nº | δC, Type | δH | δC, Type | δH | δC, Type | δH |
| 1 | 67.0, CH2 | 3.43; 3.48 | 67.1, CH2 | 3.43; 3.47 | 67.1, CH2 | 3.43; 3.48 |
| 2 | 73.0, CH | 3.58 | 73.1, CH | 3.59 | 73.1, CH | 3.59 |
| 3 | 34.2, CH2 | 1.38; 1.54 | 34.3, CH2 | 1.37; 1.54 | 34.3, CH2 | 1.38; 1.54 |
| 4 | 22.6, CH2 | 1.38; 1.62 | 22.6, CH2 | 1.38; 1.61 | 22.6, CH2 | 1.38; 1.61 |
| 5 | 38.2, CH2 | 1.40; 1.50 | 38.2, CH2 | 1.40; 1.50 | 38.1, CH2 | 1.40; 1.50 |
| 6 | 72.0, CH | 3.54 | 72.1, CH | 3.54 | 72.0, CH | 3.56 |
| 7 | 38.2, CH2 | 1.40; 1.50 | 38.2, CH2 | 1.40; 1.50 | 38.1, CH2 | 1.40; 1.50 |
| 8 | 22.6, CH2 | 1.38; 1.62 | 22.6, CH2 | 1.38; 1.61 | 22.6, CH2 | 1.38; 1.62 |
| 9 | 37.6, CH2 | 1.40; 1.52 | 37.6, CH2 | 1.39; 1.52 | 37.7, CH2 | 1.40; 1.52 |
| 10 | 71.9, CH | 3.58 | 72.2, CH | 3.58 | 72.4, CH | 3.59 |
| 11 | 41.2, CH2 | 2.20 (2H) | 41.4, CH2 | 2.20 (2H) | 41.2, CH2 | 2.19 (2H) |
| 12 | 128.6, CH | 5.69 | 128.6, CH | 5.68 | 128.5, CH | 5.70 |
| 13 | 136.0, CH | 5.53 | 135.9, CH | 5.53 | 135.9, CH | 5.55 |
| 14 | 73.2, CH | 4.05 | 73.3, CH | 4.05 | 73.2, CH | 4.05 |
| 15 | 41.7, CH2 | 2.25 (2H) | 41.8, CH2 | 2.24 (2H) | 41.7, CH2 | 2.24 (2H) |
| 16 | 129.7, CH | 5.54 | 129.6, CH | 5.53 | 129.6, CH | 5.55 |
| 17 | 137.3, CH | 5.60 | 130.1, CH | 5.60 | 130.1, CH | 5.60 |
| 18 | 37.7, CH2 | 2.08; 2.48 | 37.7, CH2 | 2.08; 2.48 | 37.7, CH2 | 2.08; 2.48 |
| 19 | 72.2, CH | 3.52 | 72.2, CH | 3.52 | 72.1, CH | 3.52 |
| 20 | 78.9, CH | 3.52 | 78.7, CH | 3.52 | 78.7, CH | 3.52 |
| 21 | 35.0, CH | 2.30 | 35.0, CH | 2.30 | 34.9, CH | 2.30 |
| 22 | 79.9, CH | 3.53 | 79.6, CH | 3.53 | 79.7, CH | 3.53 |
| 23 | 71.7, CH | 3.71 | 71.2, CH | 3.71 | 71.7, CH | 3.72 |
| 24 | 40.7, CH2 | 1.54; 1.91 | 40.9, CH2 | 1.53; 1.91 | 40.8, CH2 | 1.54; 1.90 |
| 25 | 71.1, CH | 3.86 | 71.1, CH | 3.86 | 70.1, CH | 3.87 |
| 26 | 36.2, CH2 | 1.59; 1.68 | 37.4, CH2 | 1.59; 1.68 | 36.2, CH2 | 1.59; 1.68 |
| 27 | 36.8, CH2 | 2.12; 2.21 | 36.5, CH2 | 2.12; 2.21 | 36.4, CH2 | 1.54; 1.90 |
| 28 | 139.0, C | 139.0, C | 139.1, C | |||
| 29 | 125.9, CH | 5.48 | 125.9, CH | 5.48 | 125.8, CH | 5.48 |
| 30 | 67.6, CH | 4.55 | 67.6, CH | 4.55 | 67.6, CH | 4.56 |
| 31 | 72.0, CH | 3.69 | 72.0, CH | 3.69 | 72.0, CH | 3.68 |
| 32 | 78.8, CH | 3.96 | 78.9, CH | 3.97 | 78.8, CH | 3.96 |
| 33 | 67.1, CH | 3.97 | 68.4, CH | 4.04 | 68.4, CH | 4.05 |
| 34 | 68.4, CH | 4.04 | 68.4, CH | 3.97 | 67.1, CH | 3.98 |
| 35 | 30.0, CH2 | 1.79 (2H) | 30.1, CH2 | 1.79 (2H) | 30.1, CH2 | 1.79 (2H) |
| 36 | 75.3, CH | 3.49 | 75.3, CH | 3.49 | 75.3, CH | 3.49 |
| 37 | 74.2, CH | 3.60 | 74.1, CH | 3.60 | 74.1, CH | 3.61 |
| 38 | 32.1, CH2 | 1.57; 1.97 | 32.3 CH2 | 1.57; 1.97 | 32.2 CH2 | 1.56; 1.97 |
| 39 | 27.8, CH2 | 2.10; 2.42 | 27.9, CH2 | 2.10; 2.42 | 28.0, CH2 | 2.10; 2.41 |
| 40 | 151.4, C | 151.1, C | 151.2, C | |||
| 41 | 76.3, CH | 4.18 | 76.2, CH | 4.18 | 76.1, CH | 4.19 |
| 42 | 74.1, CH | 3.35 | 75.0, CH | 3.34 | 75.0, CH | 3.35 |
| 43 | 70.0, CH | 4.05 | 70.1, CH | 4.04 | 70.2, CH | 4.04 |
| 44 | 31.1 CH2 | 1.56; 2.09 | 31.3, CH2 | 1.56; 2.09 | 31.2, CH2 | 1.56; 2.09 |
| 45 | 66.8, CH | 4.05 | 67.1, CH | 4.05 | 67.2, CH | 4.05 |
| 46 | 68.4, CH | 4.05 | 68.4, CH | 4.04 | 68.4, CH | 4.05 |
| 47 | 80.2, CH | 3.74 | 80.3, CH | 3.75 | 80.1, CH | 3.75 |
| 48 | 71.6, CH | 3.97 | 71.7, CH | 3.96 | 71.6, CH | 3.97 |
| 49 | 73.8, CH | 4.37 | 73.9, CH | 4.36 | 73.7, CH | 4.37 |
| 50 | 128.6, CH | 5.64 | 128.6, CH | 5.63 | 128.5, CH | 5.66 |
| 51 | 134.9, CH | 5.80 | 135.0, CH | 5.80 | 134.7, CH | 5.83 |
| 52 | 29.3, CH2 | 2.16 (2H) | 29.4, CH2 | 2.15 (2H) | 29.4, CH2 | 2.18 (2H) |
| 53 | 37.6, CH2 | 1.60; 1.64 | 37.6, CH2 | 1.62 (2H) | 38.8 *, CH2 | 2.16 * (2H) |
| 54 | 72.2, CH | 4.12 | 72.4, CH | 4.11 | 182.8 *, C | |
| 55 | 137.0, CH | 5.69 | 133.8, CH | 5.67 | 6.7, CH3 | 0.97 |
| 56 | 130.7, CH | 6.23 | 130.7, CH | 6.23 | 17.1, CH3 | 1.75 |
| 57 | 130.7, CH | 6.23 | 130.7, CH | 6.23 | 112.6, CH2 | 4.99; 5.09 |
| 58 | 137.0, CH | 5.69 | 133.8, CH | 5.67 | ||
| 59 | 72.8, CH | 4.10 | 72.4, CH | 4.11 | ||
| 60 | 34.2, CH2 | 1.59; 1.71 | 33.7, CH2 | 1.71; 1.73 | ||
| 61 | 34.2, CH2 | 1.38; 1.54 | 34.2, CH2 | 1.71; 1.87 | ||
| 62 | 73.0, CH | 3.58 | 77.3, CH | 4.50 | ||
| 63 | 67.8, CH2 | 3.43; 3.48 | 69.1, CH2 | 4.10; 4.26 | ||
| 64 | 6.6, CH3 | 0.98 | 6.7, CH3 | 0.98 | ||
| 65 | 17.1, CH3 | 1.75 | 17.1, CH3 | 1.75 | ||
| 66 | 112.8, CH2 | 4.99; 5.08 | 112.7, CH2 | 4.99; 5.08 | ||
| Compound | CLog P | Molecular Fragment |
|---|---|---|
| AM3 | 4.32 |  |
| Luteophanol D | 0.44 |  |
| AM20B | −1.23 |  |
| AM24 | −2.73 |  |
| AM25 | −2.82 |  |
| AM26 | −1.20 |  |
| AM27 | −1.17 |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-Amador, A.; Molina-Miras, A.; López-Rosales, L.; Sánchez-Mirón, A.; García-Camacho, F.; Souto, M.L.; Fernández, J.J. Isolation and Structural Elucidation of New Amphidinol Analogues from Amphidinium carterae Cultivated in a Pilot-Scale Photobioreactor. Mar. Drugs 2021, 19, 432. https://doi.org/10.3390/md19080432
Morales-Amador A, Molina-Miras A, López-Rosales L, Sánchez-Mirón A, García-Camacho F, Souto ML, Fernández JJ. Isolation and Structural Elucidation of New Amphidinol Analogues from Amphidinium carterae Cultivated in a Pilot-Scale Photobioreactor. Marine Drugs. 2021; 19(8):432. https://doi.org/10.3390/md19080432
Chicago/Turabian StyleMorales-Amador, Adrián, Alejandro Molina-Miras, Lorenzo López-Rosales, Asterio Sánchez-Mirón, Francisco García-Camacho, María L. Souto, and José J. Fernández. 2021. "Isolation and Structural Elucidation of New Amphidinol Analogues from Amphidinium carterae Cultivated in a Pilot-Scale Photobioreactor" Marine Drugs 19, no. 8: 432. https://doi.org/10.3390/md19080432