Comparison of Biochemical Characteristics, Action Models, and Enzymatic Mechanisms of a Novel Exolytic and Two Endolytic Lyases with Mannuronate Preference
Abstract
:1. Introduction
2. Results
2.1. Sequence Characteristics of the Alginate Lyases
2.2. Production and Purification of the Full-Length and Truncated Recombinant Proteins
2.3. Enzyme Characteristics of the Recombinant Proteins
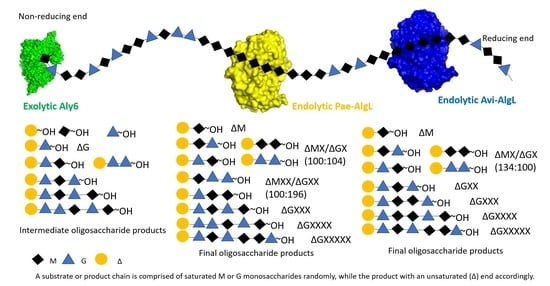
2.4. Degradation Patterns
2.5. Catalytic Mechanism of Aly6
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Carbohydrates, and Growth Conditions
4.2. Gene and Protein Sequences
4.3. Construction of Expression Vectors
4.4. Heterologous Expression and Purification of Recombinant Proteins
4.5. Enzyme Activity Assays
4.6. Biochemical Characterization of Recombinant Proteins
4.7. Comparison of Polysaccharide-Degrading Patterns
4.8. Comparison of Oligosaccharide Degradation Patterns
4.9. Analysis of the Catalytic Mechanism of the Recombinant Proteins
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Meillisa, A.; Woo, H.C.; Chun, B.S. Production of Monosaccharides and Bio-active Compounds Derived from Marine Polysaccharides using Subcritical Water Hydrolysis. Food Chem. 2015, 171, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.N.; Edgar, K.J. Alginate Derivatization: A Review of Chemistry, Properties and Applications. Biomaterials 2012, 33, 3279–3305. [Google Scholar] [CrossRef]
- Zhu, B.W.; Yin, H. Alginate Lyase: Review of Major Sources and Classification, Properties, Structure-function Analysis and Applications. Bioengineered 2015, 6, 125–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.S.; Lee, C.G.; Lee, E.Y. Alginate Lyase: Structure, Property, and Application. Biotechnol. Bioprocess Eng. 2011, 16, 843–851. [Google Scholar] [CrossRef]
- Papageorgiou, S.K.; Katsaros, F.K.; Kouvelos, E.P.; Nolan, J.W.; Deit, H.L.; Kanellopoulos, N.K. Heavy Metal Sorption by Calcium Alginate Beads from Laminaria Digitata. J. Hazard. Mater. 2006, 137, 1765–1772. [Google Scholar] [CrossRef] [PubMed]
- Ensor, S.A.; Sofos, J.N.; Schmidt, G.R. Optimization of Algin/Calcium Binder in Restructured Beef. J. Muscle Foods 1990, 1, 197–206. [Google Scholar] [CrossRef]
- Raybaudi-Massilia, R.M.; Mosqueda-Melgar, J.; Martin-Belloso, O. Edible Alginate-based Coating as Carrier of Antimicrobials to Improve Shelf-life and Safety of Fresh-cut Melon. Int. J. Food Microbiol. 2008, 121, 313–327. [Google Scholar] [CrossRef]
- Sen, F.; Uzunsoy, I.; Basturk, E.; Kahraman, M.V. Antimicrobial Agent-free Hybrid Cationic Starch/Sodium Alginate Polyelectrolyte Films for Food Packaging Materials. Carbohydr. Poly-Mers 2017, 170, 264–270. [Google Scholar] [CrossRef]
- Narayanan, K.B.; Han, S.S. Dual-crosslinked Poly (Vinyl Alcohol)/Sodium Alginate/Silver Nanocomposite Beads-A Promising Antimicrobial Material. Food Chem. 2017, 234, 103–110. [Google Scholar] [CrossRef]
- Guo, W.; Feng, J.; Geng, W.; Song, C.; Wang, Y.; Ning, C.; Wang, S. Augmented Production of Alginate Oligosaccharides by the Pseudomonas mendocina NK-01 Mutant. Carbohydr. Res. 2012, 352, 109–116. [Google Scholar] [CrossRef]
- Li, S.; He, N.; Wan, L. Efficiently Anti-Obesity Effects of Unsaturated Alginate Oligosaccharides (UAOS) in High-Fat Diet (HFD)-Fed Mice. Mar. Drugs 2019, 17, 540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, S.; Chen, X.; Dai, Z. Biological Activity of Alginate Oligosaccharides and Its Application in Food. Hans J. Agric. Sci. 2019, 9, 195–199. [Google Scholar]
- Zhou, R.; Shi, X.; Gao, Y.; Cai, N.; Jiang, Z.D.; Xu, X. Anti-inflammatory Activity of Guluronate Oligosaccharides Obtained by Oxidative Degradation from Alginate in Lipopolysaccharide-Activated Murine Macrophage RAW 264.7 Cells. J. Agric. Food Chem. 2015, 63, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Geng, M.; Li, J.; Xin, X.; Wang, J.; Tang, M.; Zhang, J.; Zhang, X.; Ding, J. Acidic Oligosaccharide Sugar Chain, a Marine-Derived Acidic Oligosaccharide, Inhibits the Cytotoxicity and Aggregation of Amyloid Beta Protein. J. Pharmacol. Sci. 2004, 95, 248–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, T.; Li, C.; Zhao, X.; Li, G.; Guan, H. Preparation and Characterization of Guluronic acid Oligosaccharides Degraded by a Rapid Microwave Irradiation Method. Carbohydr. Res. 2013, 373, 53–58. [Google Scholar] [CrossRef]
- Aida, T.M.; Yamagata, T.; Watanabe, M.; Smith, R.L. Depoly-Merization of Sodium Alginate under Hydrothermal Conditions. Carbohydr. Poly-Mers 2010, 80, 296–302. [Google Scholar] [CrossRef]
- Chandia, N.P.; Matsuhiro, B.; Vásquez, A.E. Alginic Acids in Lessonia Aaroseate: Characterization by Formic Acid Hydrolysis and FT-IR Spectroscopy. Carbohydr. Poly-Mers 2001, 46, 81–87. [Google Scholar] [CrossRef]
- Nagasawa, N.; Mitomo, H.; Yoshii, F.; Kume, T. Radiation-Induced Degradation of Sodium Alginate. Poly-Mer Degrad. Stab. 2000, 69, 279–285. [Google Scholar] [CrossRef]
- Shimokawa, T.; Yoshida, S.; Kusakabe, I.; Takeuchi, T.; Murata, K.; Kobayashi, H. Some Properties and Action Mode of (1->4)-alpha-L-guluronan Lyase from Enterobacter cloacae M-1. Carbohyd. Res. 1997, 304, 125–132. [Google Scholar] [CrossRef]
- Oliveira, R.L.; Silva, M.F.; Silva, S.P.; Araújo, A.C.V.; Cavalcanti, J.V.F.L.; Converti, A.; Portob, T.S. Fructo-Oligosaccharides Production by an Aspergillus aculeatus Commercial Enzyme Preparation with Fructosyltransferase Activity Covalently Immobilized on Fe3O4–Chitosan-Magnetic Nanoparticles. Int. J. Biol. Macromol. 2020, 150, 922–929. [Google Scholar] [CrossRef]
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. Alginate Lyase: Review of Major Sources and Enzyme Characteristics, Structure-Function Analysis, Biological Roles, and Applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Wefers, D. Chromatographic Analysis of Alginate Degradation by Five Recombinant Alginate Lyases from Cellulophaga algicola DSM 14237. Food Chem. 2019, 299, 125142. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.Y.; Wang, Q.Z.; Lu, M.Q.; Chao, X.; Li, F.; Zhang, R.; Wei, L.; Huang, S. AlgM4: A New Salt-Activated Alginate Lyase of the PL7 Family with Endolytic Activity. Mar. Drugs 2018, 16, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Li, N.N.; Yang, S.X.; Yu, Y.; Han, Z.L.; Li, L.; Mou, H.J. Study on Expression and Action Mode of Recombinant Alginate Lyases Based on Conserved Domains Reconstruction. Appl. Microbiol. Biotechnol. 2019, 103, 807–817. [Google Scholar] [CrossRef]
- Hobbs, J.K.; Lee, S.M.; Robb, M.; Hof, F.; Barr, C.; Abe, K.T.; Hehemann, J.H.; Mclean, R.; Abbott, D.W.; Boraston, A.B. KdgF, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate. Proc. Natl. Acad. Sci. USA 2016, 113, 6188–6193. [Google Scholar] [CrossRef] [Green Version]
- Enquist-Newman, M.; Faust, A.M.E.; Bravo, D.D.; Santos, C.N.S.; Raisner, R.M.; Hanel, A.; Sarvabhowman, P.; Le, C.; Regitsky, D.D.; Cooper, S.R.; et al. Efficient Ethanol Production from Brown Macroalgae Sugars by a Synthetic Yeast Platform. Nature 2014, 505, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Sasaki, Y.; Motone, K.; Shibata, T.; Tanaka, R.; Miyake, H.; Mori, T.; Kuroda, K.; Ueda, M. Construction of Bioengineered Yeast Platform for Direct Bioethanol Production from Alginate and Mannitol. Appl. Microbiol. Biotechnol. 2017, 101, 6627–6636. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.I.; Choi, S.H.; Lee, E.Y.; Kim, H.S. Molecular Cloning, Purification, and Characterization of a Novel Poly MG-specific Alginate Lyase Responsible for Alginate MG Block Degradation in Stenotrophomas maltopholia KJ-2. Appl. Microbiol. Biotechnol. 2012, 95, 1643–1653. [Google Scholar] [CrossRef] [PubMed]
- Han, W.J.; Gu, J.Y.; Cheng, Y.Y.; Liu, H.H.; Li, Y.Z.; Li, F.C. Novel Alginate Lyase (Aly5) from a Polysaccharide-degrading Marine Bacterium, Flammeovirga sp. strain MY04: Effects of Module Truncation on Biochemical Characteristics, Alginate Degradation Patterns, and Oligosaccharide-Yielding Properties. Appl. Environ. Microbiol. 2016, 82, 364–374. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.Y.; Wang, D.D.; Gu, J.Y.; Li, J.G.; Liu, H.H.; Li, F.C.; Han, W.J. Biochemical Characteristics and Variable Alginate-Degrading Modes of a Novel Bifunctional Endolytic Alginate Lyase. Appl. Environ. Microbiol. 2017, 83, e01608-17. [Google Scholar] [CrossRef] [Green Version]
- Peng, C.E.; Wang, Q.B.; Lu, D.R.; Han, W.J.; Li, F.C. A Novel Bifunctional Endolytic Alginate Lyase with Variable Action Modes and Versatile Monosaccharide-Yielding Properties. Front. Microbiol. 2018, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.W.; Zhang, Z.; Liu, Y.; Gu, J.Y.; Cheng, Y.Y.; Hu, W.; Li, Y.Z.; Han, W.J. The Second Chromosome Promotes the Adaptation of the Genus Flammeovirga to Complex Environments. Microbiol. Spectr. 2021, 9, e00980-21. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yu, Y.; Yang, S.X.; Shi, X.H.; Mou, H.J.; Li, L. Expression and Characterization of a New PolyG-Specific Alginate Lyase from Marine Bacterium Microbulbifer sp. Q7. Front Microbiol. 2018, 9, 2894. [Google Scholar] [CrossRef]
- Wang, Y.N.; Chen, X.H.; Bi, X.L.; Ren, X.L.; Zhou, Y.; Han, Y.T.; Yao, R.Y.; Li, S.Y. Characterization of an Alkaline Alginate Lyase with pH-Stable and Thermo-Tolerance Property. Mar. Drugs 2018, 17, 308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravanal, M.C.; Pezoa-Conte, R.; von Schoultz, S.; Hemming, J.; Salazar, O.; Anugwom, I.; Jogunola, O.; Mäki-Arvel, P.; Willför, S.; Mikkola María, J.P.; et al. Comparison of Different Types of Pretreatment and Enzymatic Saccharification of Macrocystis pyrifera for the Production of Biofuel. Algal Res. 2016, 13, 141–147. [Google Scholar] [CrossRef]
- Perrin, B.; Tyler, R.; Patrick, J.M.; Kitova, E.N.; Walvoort, M.T.C.; Little, D.J.; Whitney, J.C.; Dawson, K.D.; Weadge, J.T.; Robinson, H. P. aeruginosa SGNH Hydrolase-like Proteins AlgJ and AlgX Have Similar Topology but Separate and Distinct roles in alginate acetylation. PLoS Pathog. 2014, 10, e1004334. [Google Scholar]
- Riley, L.M.; Weadge, J.T.; Raker, P.; Baker, P.; Robinson, H.; Codee, J.D.C.; Tipton, P.A.; Ohman, D.E.; Howell, P.L. Structural and functional characterization of Pseudomonas aeruginosa AlgX: Role of AlgX in alginate acetylation. J. Bio. Chem. 2013, 288, 22299–22314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, I.D.; Rehman, Z.U.; Moradali, M.F.; Wang, Y.J.; Rehm, B.H.A. Microbial alginate production, modification and its applications. Microb. Biotechnol. 2013, 6, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrell, E.K.; Tipton, P.A. Functional Characterization of AlgL, an Alginate Lyase from Pseudomonas aeruginosa. Biothemistry 2012, 51, 10259–10266. [Google Scholar] [CrossRef] [Green Version]
- Belik, A.; Silchenko, A.; Malyarenko, O.; Rasin, A.; Kiseleva, M.; Kusaykin, M.; Ermakova, S. Two New Alginate Lyases of PL7 and PL6 Families from Polysaccharide-Degrading Bacterium Formosa algae KMM 3553T: Structure, Properties, and Products Analysis. Mar. Drugs 2020, 18, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraiwattanapong, J.; Tsuruga, H.; Ooi, T.; Kinoshita, S. Cloning and sequencing of a Deleya marina gene encoding for alginate lyase. Biotechnol. Lett. 1999, 21, 169–174. [Google Scholar] [CrossRef]
- Ertesvåg, H.; Erlien, F.; Skjåk-Braek, G.; Bernd, R.; Valla, A.H. Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase. J. Bacteriol. 1998, 180, 3779–3784. [Google Scholar] [CrossRef] [Green Version]
- Jang, C.H.; Piao, Y.L.; Huang, X.Q.; Yoon, E.J.; Cho, H. Modeling and Re-Engineering of Azotobacter vinelandii Alginate Lyase to Enhance Its Catalytic Efficiency for Accelerating Biofilm Degradation. PLoS ONE 2016, 11, e6156197. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.; Lundqvist, L.C.E.; Jam, M.; Jeudy, A.; Barbeyrou, T.; Sandstrom, C.; Michel, G.; Czjzek, A. Comparative Characterization of Two Marine Alginate Lyases from Zobellia galactanivorans Reveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate. J. Biol. Chem. 2013, 288, 23021–23037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jun, Y.; Peng, C.; Yan, Z.; Men, Y.; Sun, Y. The Characterization and Modification of a Novel Bifunctional and Robust Alginate Lyase Derived from Marinimicrobium sp. H1. Mar. Drugs 2019, 17, 545. [Google Scholar]
- Lu, D.R.; Zhang, Q.D.; Wang, S.M.; Guan, J.W.; Jiao, R.M.; Han, N.H.; Han, W.J.; Li, F.C. Biochemical Characteristics and Synergistic Effect of Two Novel Alginate Lyases from Photobacterium sp. FC615. Biotechnol. Biofuels 2019, 12, 260. [Google Scholar] [CrossRef] [PubMed]
- Han, W.J.; Gu, J.Y.; Yan, Q.; Li, J.G.; Wu, Z.H.; Gu, Q.Q.; Li, Y.Z. A Polysaccharide-Degrading Marine Bacterium Flammeovirga sp. MY04 and Its Extracellular Garose system. J. Ocean. Univ. China 2012, 11, 375–382. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; pp. A8.40–A8.47. [Google Scholar]






| Enzymes | Alginate | Poly-M | Poly-G |
|---|---|---|---|
| rAly6 | 726 ± 2.2 | 525 ± 3.5 | 196 ± 2.9 |
| rTF-Aly6 | 692 ± 2.7 | 547.9 ± 3.3 | 183 ± 3.1 |
| rTF-Aly6-Lmodule | 34.6 ± 5.1 | 25.1 ± 4.2 | 5 ± 4.2 |
| rTF-Aly6-HPmodule | 48 ± 4.9 | 19.5 ± 6.4 | 8 ± 6.1 |
| Pae-rAlgL | 2685 ± 3.6 | 5704 ± 4.5 | 144 ± 3.3 |
| Avi-rAlgL | 4219 ± 1.8 | 8085 ± 5.9 | 98 ± 5.2 |
| Test Substrate | Product (s) | Degradation Ratio |
|---|---|---|
| ΔG | Δ | 5% |
| ΔM | Δ | 90% |
| GG | --- | --- |
| MM | --- | --- |
| ΔGX/ΔMX | UDP2 | 60% |
| ΔGX | UDP2 | 5% |
| ΔGXX | UDP3 and UDP2 | 95% |
| ΔGXXX | UDP3 and UDP2 | 98% |
| Enzymes | Mutants | Activity (U/mg) |
|---|---|---|
| Metal ions | H426A | 0 |
| Extra peptide | G224-A248 | 367 ± 2.2 |
| NNHSYW200 | W200A | 144 ± 3.1 |
| Active site residues | H197A | 46 ± 4.6 |
| Y231A | 566 ± 1.9 | |
| Y269A | 9 ± 5.4 | |
| N144A | 16 ± 4.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, L.; Li, J.; Cheng, Y.; Wang, D.; Gu, J.; Li, F.; Han, W. Comparison of Biochemical Characteristics, Action Models, and Enzymatic Mechanisms of a Novel Exolytic and Two Endolytic Lyases with Mannuronate Preference. Mar. Drugs 2021, 19, 706. https://doi.org/10.3390/md19120706
Zeng L, Li J, Cheng Y, Wang D, Gu J, Li F, Han W. Comparison of Biochemical Characteristics, Action Models, and Enzymatic Mechanisms of a Novel Exolytic and Two Endolytic Lyases with Mannuronate Preference. Marine Drugs. 2021; 19(12):706. https://doi.org/10.3390/md19120706
Chicago/Turabian StyleZeng, Lianghuan, Junge Li, Yuanyuan Cheng, Dandan Wang, Jingyan Gu, Fuchuan Li, and Wenjun Han. 2021. "Comparison of Biochemical Characteristics, Action Models, and Enzymatic Mechanisms of a Novel Exolytic and Two Endolytic Lyases with Mannuronate Preference" Marine Drugs 19, no. 12: 706. https://doi.org/10.3390/md19120706