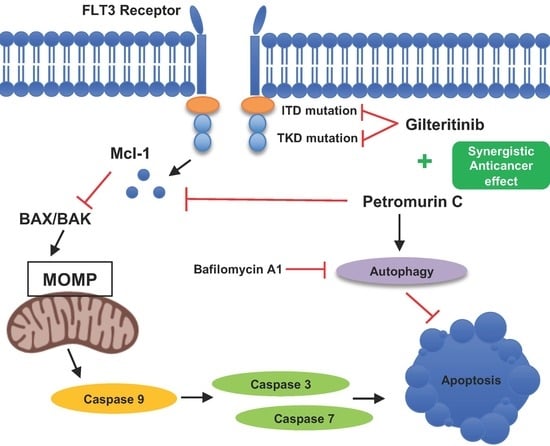
Petromurin C Induces Protective Autophagy and Apoptosis in FLT3-ITD-Positive AML: Synergy with Gilteritinib
Abstract
:1. Introduction
2. Results
2.1. Chemical Structure and Druggability of Petromurin C
2.2. Cytostatic and Cytotoxic Activities of Petromurin C
2.3. Petromurin C Induces Cell Death
2.4. Petromurin C Activates the Mitochondrial Cell Death Pathway via Inhibition of Mcl-1 and Activation of Pro-Caspases 3/7 and 9
2.5. Petromurin C Triggers Formation of Cytoplasmic Vesicles
2.6. Petromurin C Triggers a Rapid, Protective Autophagic Flux
2.7. Petromurin C Generates Mitochondrial Membrane Potential Loss and Mitochondrial Stress
2.8. Synergistic Effects of a Combination of Petromurin C and Gilteritinib on Cell Death
2.9. Investigation of Morphological Changes after Combination Treatments
3. Discussion
4. Materials and Methods
4.1. Compound Isolation
Sponge Collection and Compound Isolation
4.2. Biological Assays
4.2.1. Chemicals
4.2.2. Cell Proliferation and Viability Assays
4.2.3. Colony Formation Assay
4.2.4. Zebrafish Toxicity Assay
4.2.5. Evaluation of Cell Death
4.2.6. Protein Extraction and Western Blots
4.2.7. Morphological Analysis
4.2.8. Transmission Electron Microscopy (TEM)
4.2.9. Analysis of the Mitochondrial Membrane Potential
4.2.10. Determination of the Oxygen Consumption Rate
4.2.11. In Silico Drug-Likeness Properties
4.2.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Khwaja, A.; Bjorkholm, M.; Gale, R.E.; Levine, R.L.; Jordan, C.T.; Ehninger, G.; Bloomfield, C.D.; Estey, E.; Burnett, A.; Cornelissen, J.J.; et al. Acute myeloid leukaemia. Nat. Rev. Dis. Prim. 2016, 2, 16010. [Google Scholar] [CrossRef]
- Thein, M.S.; Ershler, W.B.; Jemal, A.; Yates, J.W.; Baer, M.R. Outcome of older patients with acute myeloid leukemia. Cancer 2013, 119, 2720–2727. [Google Scholar] [CrossRef] [PubMed]
- Stirewalt, D.L.; Radich, J.P. The role of FLT3 in haematopoietic malignancies. Nat. Rev. Cancer 2003, 3, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Perl, A.E.; Altman, J.K.; Cortes, J.; Smith, C.; Litzow, M.; Baer, M.R.; Claxton, D.; Erba, H.P.; Gill, S.; Goldberg, S.; et al. Selective inhibition of FLT3 by gilteritinib in relapsed or refractory acute myeloid leukaemia: A multicentre, first-in-human, open-label, phase 1–2 study. Lancet Oncol. 2017, 18, 1061–1075. [Google Scholar] [CrossRef]
- Florean, C.; Kim, R.K.; Schnekenburger, M.; Kim, H.-J.; Moriou, C.; Debitus, C.; Dicato, M.; Al-Mourabit, A.; Han, W.B.; Diederich, M. Synergistic AML cell death induction by marine cytotoxin (+)-1(R), 6(S), 1′(R), 6′(S), 11(R), 17(S)-Fistularin-3 and Bcl-2 inhibitor venetoclax. Mar. Drug. 2018, 16, 518. [Google Scholar] [CrossRef] [Green Version]
- Buttachon, S.; Ramos, A.; Inácio, Â.; Dethoup, T.; Gales, L.; Lee, M.; Costa, P.; Silva, A.; Sekeroglu, N.; Rocha, E.; et al. Bis-Indolyl benzenoids, Hydroxypyrrolidine derivatives and other constituents from cultures of the marine sponge-associated fungus Aspergillus candidus KUFA0062. Mar. Drug. 2018, 16, 119. [Google Scholar] [CrossRef] [Green Version]
- Radogna, F.; Dicato, M.; Diederich, M. Cancer-type-specific crosstalk between autophagy, necroptosis and apoptosis as a pharmacological target. Biochem. Pharmacol. 2015, 94, 1–11. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the nomenclature Committee on cell death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Gordy, C.; He, Y.-W. The crosstalk between autophagy and apoptosis: Where does this lead? Protein Cell 2012, 3, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Short, N.J.; Kantarjian, H.; Ravandi, F.; Daver, N. Emerging treatment paradigms with FLT3 inhibitors in acute myeloid leukemia. Ther. Adv. Hematol. 2019, 10, 2040620719827310. [Google Scholar] [CrossRef] [Green Version]
- Ueno, Y.; Mori, M.; Kamiyama, Y.; Saito, R.; Kaneko, N.; Isshiki, E.; Kuromitsu, S.; Takeuchi, M. Evaluation of gilteritinib in combination with chemotherapy in preclinical models of FLT3-ITD+ acute myeloid leukemia. Oncotarget 2019, 10, 2530–2545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carles, F.; Bourg, S.; Meyer, C.; Bonnet, P. PKIDB: A curated, annotated and updated database of protein kinase inhibitors in clinical trials. Molecules 2018, 23, 908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerella, C.; Muller, F.; Gaigneaux, A.; Radogna, F.; Viry, E.; Chateauvieux, S.; Dicato, M.; Diederich, M. Early downregulation of Mcl-1 regulates apoptosis triggered by cardiac glycoside UNBS1450. Cell Death Dis. 2015, 6, e1782. [Google Scholar] [CrossRef] [PubMed]
- Chaitanya, G.V.; Steven, A.J.; Babu, P.P. PARP-1 cleavage fragments: Signatures of cell-death proteases in neurodegeneration. Cell Commun. Signal 2010, 8, 31. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.; Majeti, R. Biology and relevance of human acute myeloid leukemia stem cells. Blood 2017, 129, 1577–1585. [Google Scholar] [CrossRef]
- Lee, L.Y.; Hernandez, D.; Rajkhowa, T.; Smith, S.C.; Raman, J.R.; Nguyen, B.; Small, D.; Levis, M. Preclinical studies of gilteritinib, a next-generation FLT3 inhibitor. Blood 2017, 129, 257–260. [Google Scholar] [CrossRef] [Green Version]
- Bohl, S.R.; Bullinger, L.; Rücker, F.G. New targeted agents in acute myeloid leukemia: New Hope on the rise. Int. J. Mol. Sci. 2019, 20, 1983. [Google Scholar] [CrossRef] [Green Version]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef]
- Watts, J.; Nimer, S. Recent advances in the understanding and treatment of acute myeloid leukemia. F1000Research 2018, 7, 1196. [Google Scholar] [CrossRef] [Green Version]
- Pereira, F. Have marine natural product drug discovery efforts been productive and how can we improve their efficiency? Expert Opin. Drug Dis. 2019, 14, 717–722. [Google Scholar] [CrossRef] [Green Version]
- Khalifa, A.M.S.; Elias, N.; Farag, A.M.; Chen, L.; Saeed, A.; Hegazy, F.M.-E.; Moustafa, S.M.; Abd El-Wahed, A.; Al-Mousawi, M.S.; Musharraf, G.S.; et al. Marine natural products: A source of novel anticancer drugs. Mar. Drug. 2019, 17, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florian, M.; Claudia, C.; Flavia, R.; Mario, D.; Marc, D. Effects of natural products on Mcl-1 expression and function. Curr. Med. Chem. 2015, 22, 3447–3461. [Google Scholar]
- Ding, Y.-S.; Kim, W.-S.; Park, S.J.; Kim, S.-K. Apoptotic effect of physcion isolated from marine fungus Microsporum sp. in PC3 human prostate cancer cells. Fish. Aquat. Sci. 2018, 21, 22. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Orsini, M.; Morceau, F.; Dicato, M.; Diederich, M. Autophagy as a pharmacological target in hematopoiesis and hematological disorders. Biochem. Pharmacol. 2018, 152, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Eskelinen, E.-L.; Deretic, V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes… wait, I’m confused. Autophagy 2014, 10, 549–551. [Google Scholar] [CrossRef] [Green Version]
- Wan, X.; Serrill, D.J.; Humphreys, R.I.; Tan, M.; McPhail, L.K.; Ganley, G.I.; Ishmael, E.J. ATG5 promotes death signaling in response to the cyclic depsipeptides Coibamide A and Apratoxin, A. Mar. Drugs 2018, 16, 77. [Google Scholar] [CrossRef] [Green Version]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and animal models: Are 3D cultures the ideal tool to study cancer-microenvironment interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [Green Version]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef]
- Melissaridou, S.; Wiechec, E.; Magan, M.; Jain, M.V.; Chung, M.K.; Farnebo, L.; Roberg, K. The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer. Cancer Cell Int. 2019, 19, 16. [Google Scholar] [CrossRef] [Green Version]
- Adcock, A.; Trivedi, G. Three-Dimensional (3D) cell cultures in cell-based assays for in-vitro evaluation of anticancer drugs. J. Anal. Bioanal. Tech. 2015, 6, 3. [Google Scholar] [CrossRef]
- Zheng, R.; Bailey, E.; Nguyen, B.; Yang, X.; Piloto, O.; Levis, M.; Small, D. Further activation of FLT3 mutants by FLT3 ligand. Oncogene 2011, 30, 4004–4014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]













| Drug-Likeness | Petromurin C |
|---|---|
| Mass (g/mol) | 444.49 |
| LogP | 5.26 |
| Hydrogen bond donors | 3 |
| Hydrogen bond acceptors | 7 |
| Cell Line | 24 h | 48 h | 72 h | |||
|---|---|---|---|---|---|---|
| IC50 (µM) | GI50 (µM) | IC50 (µM) | GI50 (µM) | IC50 (µM) | GI50 (µM) | |
| MV4-11 | >50 | >50 | >50 | 18.56 ± 3.61 | 43.14 ± 2.88 | 19.28 ± 3.66 |
| U937 | >50 | 31.68 ± 6.83 | >50 | 10.15 ± 3.42 | >50 | 11.55 ± 2.34 |
| Petromurin C (µM) | Gilteritinib (nM) | Effect (Fa) | CI |
|---|---|---|---|
| 5 | 100 | 0.26 | 0.31 |
| 5 | 250 | 0.39 | 0.12 |
| 5 | 500 | 0.38 | 0.22 |
| 30 | 100 | 0.26 | 0.90 |
| 30 | 250 | 0.56 | 0.15 |
| 30 | 500 | 0.48 | 0.24 |
| 50 | 100 | 0.60 | 0.20 |
| 50 | 250 | 0.65 | 0.15 |
| 50 | 500 | 0.62 | 0.18 |
| Petromurin C (µM) | Gilteritinib (nM) | Effect (Fa) | CI |
|---|---|---|---|
| 10 | 0.5 | 0.42 | 0.59 |
| 10 | 1.0 | 0.52 | 0.81 |
| 10 | 2.0 | 0.84 | 0.55 |
| 30 | 0.5 | 0.59 | 0.43 |
| 30 | 1.0 | 0.65 | 0.61 |
| 30 | 2.0 | 0.85 | 0.54 |
| 50 | 0.5 | 0.60 | 0.48 |
| 50 | 1.0 | 0.69 | 0.58 |
| 50 | 2.0 | 0.89 | 0.43 |
| Vacuole Size/Area (µm2) | Petromurin C (30 µM) | Petromurin C (30 µM) and Gilteritinib (250 nM) |
|---|---|---|
| Area > 0.2 µm2 | 27 | 22 |
| Area < 0.2 µm2 | 37 | 154 |
| All | 64 | 175 |
| Area > 0.2 µm2 (%) | 42.2 | 12.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, Y.N.; Song, S.; Orlikova-Boyer, B.; Cerella, C.; Christov, C.; Kijjoa, A.; Diederich, M. Petromurin C Induces Protective Autophagy and Apoptosis in FLT3-ITD-Positive AML: Synergy with Gilteritinib. Mar. Drugs 2020, 18, 57. https://doi.org/10.3390/md18010057
Ha YN, Song S, Orlikova-Boyer B, Cerella C, Christov C, Kijjoa A, Diederich M. Petromurin C Induces Protective Autophagy and Apoptosis in FLT3-ITD-Positive AML: Synergy with Gilteritinib. Marine Drugs. 2020; 18(1):57. https://doi.org/10.3390/md18010057
Chicago/Turabian StyleHa, You Na, Sungmi Song, Barbora Orlikova-Boyer, Claudia Cerella, Christo Christov, Anake Kijjoa, and Marc Diederich. 2020. "Petromurin C Induces Protective Autophagy and Apoptosis in FLT3-ITD-Positive AML: Synergy with Gilteritinib" Marine Drugs 18, no. 1: 57. https://doi.org/10.3390/md18010057