The Use of CBD and Its Synthetic Analog HU308 in HIV-1-Infected Myeloid Cells
Abstract
:1. Introduction
2. Results
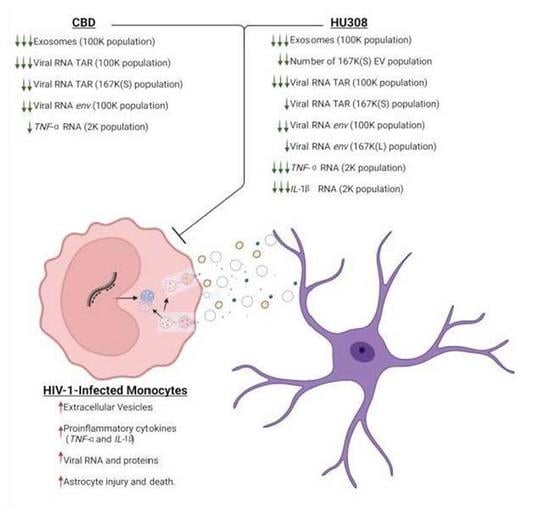
2.1. CBD Decreases the Classical Exosome EV Subpopulation
2.2. CBD Decreases Viral RNA and Proinflammatory Cytokine mRNA
2.3. Analog of CBD (HU308) Similarly Reduces EV Subpopulations
2.4. HU308 Reduces Both Viral and Proinflammatory RNAs
2.5. Decrease in Viral RNA in Primary Macrophages Treated with HU308 or CBD
2.6. Effect of HU308 and CBD in a 3D Mini-Brain Infection Model
2.7. HU308 Treatment Reduces Viral RNA in Humanized Mice
3. Discussion
4. Materials and Methods
4.1. Cells and Reagents
4.2. Primary Cells
4.3. Serum EV-Depleted Medium
4.4. ZetaView Nanoparticle Tracking Analysis (NTA)
4.5. EV Isolation via Differential Ultracentrifugation
4.6. RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR (RT-qPCR)
4.7. Preparation of Whole Cell Extracts
4.8. Western Blot
4.9. Cell Viability Assay
4.10. Neurosphere 3D Model
4.11. Humanized Mice
4.12. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- HIV and AIDS. Available online: https://www.who.int/news-room/fact-sheets/detail/hiv-aids (accessed on 29 June 2023).
- Rojas-Celis, V.; Valiente-Echeverría, F.; Soto-Rifo, R.; Toro-Ascuy, D. New Challenges of HIV-1 Infection: How HIV-1 Attacks and Resides in the Central Nervous System. Cells 2019, 8, 1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costiniuk, C.T.; Jenabian, M.-A. Cannabinoids and Inflammation: Implications for People Living with HIV. AIDS Lond. Engl. 2019, 33, 2273–2288. [Google Scholar] [CrossRef] [PubMed]
- DeMarino, C.; Cowen, M.; Khatkar, P.; Cotto, B.; Branscome, H.; Kim, Y.; Sharif, S.A.; Agbottah, E.T.; Zhou, W.; Costiniuk, C.T.; et al. Cannabinoids Reduce Extracellular Vesicle Release from HIV-1 Infected Myeloid Cells and Inhibit Viral Transcription. Cells 2022, 11, 723. [Google Scholar] [CrossRef]
- Bhatti, A.B.; Usman, M.; Kandi, V. Current Scenario of HIV/AIDS, Treatment Options, and Major Challenges with Compliance to Antiretroviral Therapy. Cureus 2016, 8, e515. [Google Scholar] [CrossRef] [Green Version]
- Saylor, D.; Dickens, A.M.; Sacktor, N.; Haughey, N.; Slusher, B.; Pletnikov, M.; Mankowski, J.L.; Brown, A.; Volsky, D.J.; McArthur, J.C. HIV-Associated Neurocognitive Disorder—Pathogenesis and Prospects for Treatment. Nat. Rev. Neurol. 2016, 12, 234–248. [Google Scholar] [CrossRef]
- Cirino, T.J.; McLaughlin, J.P. Mini Review: Promotion of Substance Abuse in HIV Patients: Biological Mediation by HIV-1 Tat Protein. Neurosci. Lett. 2021, 753, 135877. [Google Scholar] [CrossRef]
- Khan, N.; Haughey, N.J.; Nath, A.; Geiger, J.D. Involvement of Organelles and Inter-Organellar Signaling in the Pathogenesis of HIV-1 Associated Neurocognitive Disorder and Alzheimer’s Disease. Brain Res. 2019, 1722, 146389. [Google Scholar] [CrossRef]
- Anzinger, J.J.; Butterfield, T.R.; Angelovich, T.A.; Crowe, S.M.; Palmer, C.S. Monocytes as Regulators of Inflammation and HIV-Related Comorbidities during CART. J. Immunol. Res. 2014, 2014, 569819. [Google Scholar] [CrossRef] [PubMed]
- Schlachetzki, J.C.M.; Zhou, Y.; Glass, C.K. Human Microglia Phenotypes in the Brain Associated with HIV Infection. Curr. Opin. Neurobiol. 2022, 77, 102637. [Google Scholar] [CrossRef]
- DeMarino, C.; Pleet, M.L.; Cowen, M.; Barclay, R.A.; Akpamagbo, Y.; Erickson, J.; Ndembi, N.; Charurat, M.; Jumare, J.; Bwala, S.; et al. Antiretroviral Drugs Alter the Content of Extracellular Vesicles from HIV-1-Infected Cells. Sci. Rep. 2018, 8, 7653. [Google Scholar] [CrossRef] [Green Version]
- Hermes, D.J.; Yadav-Samudrala, B.J.; Xu, C.; Paniccia, J.E.; Meeker, R.B.; Armstrong, M.L.; Reisdorph, N.; Cravatt, B.F.; Mackie, K.; Lichtman, A.H.; et al. GPR18 Drives FAAH Inhibition-Induced Neuroprotection against HIV-1 Tat-Induced Neurodegeneration. Exp. Neurol. 2021, 341, 113699. [Google Scholar] [CrossRef] [PubMed]
- Barclay, R.A.; Schwab, A.; DeMarino, C.; Akpamagbo, Y.; Lepene, B.; Kassaye, S.; Iordanskiy, S.; Kashanchi, F. Exosomes from Uninfected Cells Activate Transcription of Latent HIV-1. J. Biol. Chem. 2017, 292, 14764. [Google Scholar] [CrossRef] [Green Version]
- Barclay, R.A.; Mensah, G.A.; Cowen, M.; DeMarino, C.; Kim, Y.; Pinto, D.O.; Erickson, J.; Kashanchi, F. Extracellular Vesicle Activation of Latent HIV-1 Is Driven by EV-Associated c-Src and Cellular SRC-1 via the PI3K/AKT/MTOR Pathway. Viruses 2020, 12, 665. [Google Scholar] [CrossRef]
- Campbell, L.A.; Mocchetti, I. Extracellular Vesicles and HIV-Associated Neurocognitive Disorders: Implications in Neuropathogenesis and Disease Diagnosis. Neurotox. Res. 2021, 39, 2098–2107. [Google Scholar] [CrossRef]
- Rahimian, P.; He, J.J. Exosome-Associated Release, Uptake, and Neurotoxicity of HIV-1 Tat Protein. J. Neurovirol. 2016, 22, 774–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubrovsky, L.; Brichacek, B.; Prashant, N.M.; Pushkarsky, T.; Mukhamedova, N.; Fleetwood, A.J.; Xu, Y.; Dragoljevic, D.; Fitzgerald, M.; Horvath, A.; et al. Extracellular Vesicles Carrying HIV-1 Nef Induce Long-Term Hyperreactivity of Myeloid Cells. Cell Rep. 2022, 41, 111674. [Google Scholar] [CrossRef]
- Sami Saribas, A.; Cicalese, S.; Ahooyi, T.M.; Khalili, K.; Amini, S.; Sariyer, I.K. HIV-1 Nef Is Released in Extracellular Vesicles Derived from Astrocytes: Evidence for Nef-Mediated Neurotoxicity. Cell Death Dis. 2018, 8, e2542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocozza, F.; Grisard, E.; Martin-Jaular, L.; Mathieu, M.; Théry, C. SnapShot: Extracellular Vesicles. Cell 2020, 182, 262. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Branscome, H.; Paul, S.; Yin, D.; El-Hage, N.; Agbottah, E.T.; Zadeh, M.A.; Liotta, L.A.; Kashanchi, F. Use of Stem Cell Extracellular Vesicles as a “Holistic” Approach to CNS Repair. Front. Cell Dev. Biol. 2020, 8, 455. [Google Scholar] [CrossRef]
- Pulliam, L.; Sun, B.; Mustapic, M.; Chawla, S.; Kapogiannis, D. Plasma Neuronal Exosomes Serve as Biomarkers of Cognitive Impairment in HIV Infection and Alzheimer’s Disease. J. Neurovirol. 2019, 25, 702–709. [Google Scholar] [CrossRef]
- Kim, Y.; Mensah, G.A.; Al Sharif, S.; Pinto, D.O.; Branscome, H.; Yelamanchili, S.V.; Cowen, M.; Erickson, J.; Khatkar, P.; Mahieux, R.; et al. Extracellular Vesicles from Infected Cells Are Released Prior to Virion Release. Cells 2021, 10, 781. [Google Scholar] [CrossRef]
- Salmond, N.; Williams, K.C. Isolation and Characterization of Extracellular Vesicles for Clinical Applications in Cancer—Time for Standardization? Nanoscale Adv. 2021, 3, 1830–1852. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.-H.; Qasim, M.; Khan, K.; Kim, J.-H. Biogenesis, Membrane Trafficking, Functions, and Next Generation Nanotherapeutics Medicine of Extracellular Vesicles. Int. J. Nanomed. 2021, 16, 3357–3383. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of Distinct Nanoparticles and Subsets of Extracellular Vesicles by Asymmetric Flow Field-Flow Fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Zijlstra, A.; Di Vizio, D. Size Matters in Nanoscale Communication. Nat. Cell Biol. 2018, 20, 228–230. [Google Scholar] [CrossRef]
- Kang, T.; Atukorala, I.; Mathivanan, S. Biogenesis of Extracellular Vesicles. In New Frontiers: Extracellular Vesicles; Mathivanan, S., Fonseka, P., Nedeva, C., Atukorala, I., Eds.; Subcellular Biochemistry; Springer International Publishing: Cham, Switzerland, 2021; pp. 19–43. ISBN 978-3-030-67171-6. [Google Scholar]
- Johnson, S.M.; Banyard, A.; Smith, C.; Mironov, A.; McCabe, M.G. Large Extracellular Vesicles Can Be Characterised by Multiplex Labelling Using Imaging Flow Cytometry. Int. J. Mol. Sci. 2020, 21, 8723. [Google Scholar] [CrossRef]
- Caobi, A.; Nair, M.; Raymond, A.D. Extracellular Vesicles in the Pathogenesis of Viral Infections in Humans. Viruses 2020, 12, 1200. [Google Scholar] [CrossRef]
- Kumar, A.; Kodidela, S.; Tadrous, E.; Cory, T.J.; Walker, C.M.; Smith, A.M.; Mukherjee, A.; Kumar, S. Extracellular Vesicles in Viral Replication and Pathogenesis and Their Potential Role in Therapeutic Intervention. Viruses 2020, 12, 887. [Google Scholar] [CrossRef]
- Chivero, E.T.; Guo, M.-L.; Periyasamy, P.; Liao, K.; Callen, S.E.; Buch, S. HIV-1 Tat Primes and Activates Microglial NLRP3 Inflammasome-Mediated Neuroinflammation. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 3599–3609. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-ΚB Signaling in Inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [Green Version]
- Watson, C.W.-M.; Paolillo, E.W.; Morgan, E.E.; Umlauf, A.; Sundermann, E.E.; Ellis, R.J.; Letendre, S.; Marcotte, T.D.; Heaton, R.K.; Grant, I. Cannabis Exposure Is Associated With a Lower Likelihood of Neurocognitive Impairment in People Living With HIV. JAIDS J. Acquir. Immune Defic. Syndr. 2020, 83, 56–64. [Google Scholar] [CrossRef]
- Watson, C.W.-M.; Sundermann, E.; Helm, J.; Paolillo, E.W.; Hong, S.; Ellis, R.J.; Letendre, S.; Marcotte, T.D.; Heaton, R.K.; Morgan, E.E.; et al. A Longitudinal Study of Cannabis Use and Risk for Cognitive and Functional Decline among Older Adults with HIV. AIDS Behav. 2023, 1–13. [Google Scholar] [CrossRef]
- Aly, E.; Masocha, W. Targeting the Endocannabinoid System for Management of HIV-Associated Neuropathic Pain: A Systematic Review. IBRO Neurosci. Rep. 2021, 10, 109–118. [Google Scholar] [CrossRef]
- Sangiovanni, E.; Fumagalli, M.; Pacchetti, B.; Piazza, S.; Magnavacca, A.; Khalilpour, S.; Melzi, G.; Martinelli, G.; Dell’Agli, M. Cannabis Sativa L. Extract and Cannabidiol Inhibit in Vitro Mediators of Skin Inflammation and Wound Injury. Phytother. Res. PTR 2019, 33, 2083–2093. [Google Scholar] [CrossRef]
- Hammell, D.C.; Zhang, L.P.; Ma, F.; Abshire, S.M.; McIlwrath, S.L.; Stinchcomb, A.L.; Westlund, K.N. Transdermal Cannabidiol Reduces Inflammation and Pain-Related Behaviours in a Rat Model of Arthritis. Eur. J. Pain Lond. Engl. 2016, 20, 936–948. [Google Scholar] [CrossRef] [Green Version]
- Philpott, H.T.; O’Brien, M.; McDougall, J.J. Attenuation of Early Phase Inflammation by Cannabidiol Prevents Pain and Nerve Damage in Rat Osteoarthritis. Pain 2017, 158, 2442–2451. [Google Scholar] [CrossRef]
- Yousaf, M.; Chang, D.; Liu, Y.; Liu, T.; Zhou, X. Neuroprotection of Cannabidiol, Its Synthetic Derivatives and Combination Preparations against Microglia-Mediated Neuroinflammation in Neurological Disorders. Molecules 2022, 27, 4961. [Google Scholar] [CrossRef]
- Burstein, S. Cannabidiol (CBD) and Its Analogs: A Review of Their Effects on Inflammation. Bioorg. Med. Chem. 2015, 23, 1377–1385. [Google Scholar] [CrossRef]
- Oláh, A.; Tóth, B.I.; Borbíró, I.; Sugawara, K.; Szöllõsi, A.G.; Czifra, G.; Pál, B.; Ambrus, L.; Kloepper, J.; Camera, E.; et al. Cannabidiol Exerts Sebostatic and Antiinflammatory Effects on Human Sebocytes. J. Clin. Investig. 2014, 124, 3713–3724. [Google Scholar] [CrossRef] [Green Version]
- Gray, R.A.; Whalley, B.J. The Proposed Mechanisms of Action of CBD in Epilepsy. Epileptic Disord. Int. Epilepsy J. Videotape 2020, 22, 10–15. [Google Scholar]
- Daneman, R.; Prat, A. The Blood–Brain Barrier. Cold Spring Harb. Perspect. Biol. 2015, 7, a020412. [Google Scholar] [CrossRef] [Green Version]
- Golub, V.; Reddy, D.S. Cannabidiol Therapy for Refractory Epilepsy and Seizure Disorders. Adv. Exp. Med. Biol. 2021, 1264, 93–110. [Google Scholar] [CrossRef]
- Vallée, A.; Vallée, J.-N.; Lecarpentier, Y. Potential Role of Cannabidiol in Parkinson’s Disease by Targeting the WNT/β-Catenin Pathway, Oxidative Stress and Inflammation. Aging 2021, 13, 10796–10813. [Google Scholar] [CrossRef]
- Fiani, B.; Sarhadi, K.J.; Soula, M.; Zafar, A.; Quadri, S.A. Current Application of Cannabidiol (CBD) in the Management and Treatment of Neurological Disorders. Neurol. Sci. Off. J. Ital. Neurol. Soc. Ital. Soc. Clin. Neurophysiol. 2020, 41, 3085–3098. [Google Scholar] [CrossRef]
- Tomita, T.; Kato, M.; Mishima, T.; Matsunaga, Y.; Sanjo, H.; Ito, K.; Minagawa, K.; Matsui, T.; Oikawa, H.; Takahashi, S.; et al. Extracellular MRNA Transported to the Nucleus Exerts Translation-Independent Function. Nat. Commun. 2021, 12, 3655. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of MRNAs and MicroRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deregibus, M.C.; Cantaluppi, V.; Calogero, R.; Lo Iacono, M.; Tetta, C.; Biancone, L.; Bruno, S.; Bussolati, B.; Camussi, G. Endothelial Progenitor Cell Derived Microvesicles Activate an Angiogenic Program in Endothelial Cells by a Horizontal Transfer of MRNA. Blood 2007, 110, 2440–2448. [Google Scholar] [CrossRef] [Green Version]
- Di Liegro, C.M.; Schiera, G.; Di Liegro, I. Extracellular Vesicle-Associated RNA as a Carrier of Epigenetic Information. Genes 2017, 8, 240. [Google Scholar] [CrossRef] [Green Version]
- Commissioner, O. FDA Regulation of Cannabis and Cannabis-Derived Products, Including Cannabidiol (CBD); FDA: Silver Spring, MD, USA, 2023. [Google Scholar]
- Arzimanoglou, A.; Brandl, U.; Cross, J.H.; Gil-Nagel, A.; Lagae, L.; Landmark, C.J.; Specchio, N.; Nabbout, R.; Thiele, E.A.; Gubbay, O.; et al. Epilepsy and Cannabidiol: A Guide to Treatment. Epileptic Disord. Int. Epilepsy J. Videotape 2020, 22, 1–14. [Google Scholar]
- Hanuš, L.; Breuer, A.; Tchilibon, S.; Shiloah, S.; Goldenberg, D.; Horowitz, M.; Pertwee, R.G.; Ross, R.A.; Mechoulam, R.; Fride, E. HU-308: A Specific Agonist for CB2, a Peripheral Cannabinoid Receptor. Proc. Natl. Acad. Sci. USA 1999, 96, 14228–14233. [Google Scholar] [CrossRef]
- Mlost, J.; Bryk, M.; Starowicz, K. Cannabidiol for Pain Treatment: Focus on Pharmacology and Mechanism of Action. Int. J. Mol. Sci. 2020, 21, 8870. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, C.; Blanchet, M.-R.; Laviolette, M.; Flamand, N. The CB2 Receptor and Its Role as a Regulator of Inflammation. Cell. Mol. Life Sci. 2016, 73, 4449–4470. [Google Scholar] [CrossRef] [Green Version]
- Szallasi, A.; Cortright, D.N.; Blum, C.A.; Eid, S.R. The Vanilloid Receptor TRPV1: 10 Years from Channel Cloning to Antagonist Proof-of-Concept. Nat. Rev. Drug Discov. 2007, 6, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Tsuzuno, T.; Mineo, S.; Yamada-Hara, M.; Aoki-Nonaka, Y.; Tabeta, K. Epithelial TRPV1 Channels: Expression, Function, and Pathogenicity in the Oral Cavity. J. Oral Biosci. 2020, 62, 235–241. [Google Scholar] [CrossRef]
- Haddad, M.; Alsalem, M.; Saleh, T.; Jaffal, S.M.; Barakat, N.A.; El-Salem, K. Interaction of the Synthetic Cannabinoid WIN55212 with Tramadol on Nociceptive Thresholds and Core Body Temperature in a Chemotherapy-Induced Peripheral Neuropathy Pain Model. Neuroreport 2023, 34, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Thapa, D.; Cairns, E.A.; Szczesniak, A.-M.; Toguri, J.T.; Caldwell, M.D.; Kelly, M.E.M. The Cannabinoids Δ8THC, CBD, and HU-308 Act via Distinct Receptors to Reduce Corneal Pain and Inflammation. Cannabis Cannabinoid Res. 2018, 3, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Henshaw, F.R.; Dewsbury, L.S.; Lim, C.K.; Steiner, G.Z. The Effects of Cannabinoids on Pro- and Anti-Inflammatory Cytokines: A Systematic Review of In Vivo Studies. Cannabis Cannabinoid Res. 2021, 6, 177–195. [Google Scholar] [CrossRef]
- Haddad, M.; Alsalem, M.; Aldossary, S.A.; Kalbouneh, H.; Jaffal, S.M.; Alshawabkeh, Q.; Al Hayek, S.; Abdelhai, O.; Barakat, N.A.; El-Salem, K. The Role of Adenosine Receptor Ligands on Inflammatory Pain: Possible Modulation of TRPV1 Receptor Function. Inflammopharmacology 2023, 31, 337–347. [Google Scholar] [CrossRef]
- Kruize, Z.; Kootstra, N.A. The Role of Macrophages in HIV-1 Persistence and Pathogenesis. Front. Microbiol. 2019, 10, 2828. [Google Scholar] [CrossRef] [Green Version]
- Branscome, H.; Khatkar, P.; Al Sharif, S.; Yin, D.; Jacob, S.; Cowen, M.; Kim, Y.; Erickson, J.; Brantner, C.A.; El-Hage, N.; et al. Retroviral Infection of Human Neurospheres and Use of Stem Cell EVs to Repair Cellular Damage. Sci. Rep. 2022, 12, 2019. [Google Scholar] [CrossRef]
- Iordanskiy, S.; Van Duyne, R.; Sampey, G.C.; Woodson, C.M.; Fry, K.; Saifuddin, M.; Guo, J.; Wu, Y.; Romerio, F.; Kashanchi, F. Therapeutic Doses of Irradiation Activate Viral Transcription and Induce Apoptosis in HIV-1 Infected Cells. Virology 2015, 485, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Sophocleous, A.; Landao-Bassonga, E.; van‘t Hof, R.J.; Idris, A.I.; Ralston, S.H. The Type 2 Cannabinoid Receptor Regulates Bone Mass and Ovariectomy-Induced Bone Loss by Affecting Osteoblast Differentiation and Bone Formation. Endocrinology 2011, 152, 2141–2149. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Jin, G.; Zhang, J.; Wei, W. Selective Activation of Cannabinoid Receptor 2 Attenuates Myocardial Infarction via Suppressing NLRP3 Inflammasome. Inflammation 2019, 42, 904–914. [Google Scholar] [CrossRef]
- Kosgodage, U.S.; Uysal-Onganer, P.; MacLatchy, A.; Mould, R.; Nunn, A.V.; Guy, G.W.; Kraev, I.; Chatterton, N.P.; Thomas, E.L.; Inal, J.M.; et al. Cannabidiol Affects Extracellular Vesicle Release, MiR21 and MiR126, and Reduces Prohibitin Protein in Glioblastoma Multiforme Cells. Transl. Oncol. 2019, 12, 513–522. [Google Scholar] [CrossRef]
- Kosgodage, U.S.; Matewele, P.; Awamaria, B.; Kraev, I.; Warde, P.; Mastroianni, G.; Nunn, A.V.; Guy, G.W.; Bell, J.D.; Inal, J.M.; et al. Cannabidiol Is a Novel Modulator of Bacterial Membrane Vesicles. Front. Cell. Infect. Microbiol. 2019, 9, 324. [Google Scholar] [CrossRef]
- Kosgodage, U.S.; Mould, R.; Henley, A.B.; Nunn, A.V.; Guy, G.W.; Thomas, E.L.; Inal, J.M.; Bell, J.D.; Lange, S. Cannabidiol (CBD) Is a Novel Inhibitor for Exosome and Microvesicle (EMV) Release in Cancer. Front. Pharmacol. 2018, 9, 889. [Google Scholar] [CrossRef] [Green Version]
- Kageyama, S.; Gudmundsson, S.R.; Sou, Y.-S.; Ichimura, Y.; Tamura, N.; Kazuno, S.; Ueno, T.; Miura, Y.; Noshiro, D.; Abe, M.; et al. P62/SQSTM1-Droplet Serves as a Platform for Autophagosome Formation and Anti-Oxidative Stress Response. Nat. Commun. 2021, 12, 16. [Google Scholar] [CrossRef]
- Tilija Pun, N.; Park, P.-H. Role of P62 in the Suppression of Inflammatory Cytokine Production by Adiponectin in Macrophages: Involvement of Autophagy and P21/Nrf2 Axis. Sci. Rep. 2017, 7, 393. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Yang, X.; Zhao, J.; Cheng, Y.; Wang, J. Mechanism and Complex Roles of HSC70 in Viral Infections. Front. Microbiol. 2020, 11, 1577. [Google Scholar] [CrossRef]
- Jiang, R.; Gao, B.; Prasad, K.; Greene, L.E.; Eisenberg, E. Hsc70 Chaperones Clathrin and Primes It to Interact with Vesicle Membranes*. J. Biol. Chem. 2000, 275, 8439–8447. [Google Scholar] [CrossRef] [Green Version]
- Taylor, L.; Christou, I.; Kapellos, T.S.; Buchan, A.; Brodermann, M.H.; Gianella-Borradori, M.; Russell, A.; Iqbal, A.J.; Greaves, D.R. Primary Macrophage Chemotaxis Induced by Cannabinoid Receptor 2 Agonists Occurs Independently of the CB2 Receptor. Sci. Rep. 2015, 5, 10682. [Google Scholar] [CrossRef]
- Mabou Tagne, A.; Pacchetti, B.; Sodergren, M.; Cosentino, M.; Marino, F. Cannabidiol for Viral Diseases: Hype or Hope? Cannabis Cannabinoid Res. 2020, 5, 121–131. [Google Scholar] [CrossRef]
- Lowe, H.I.C.; Toyang, N.J.; McLaughlin, W. Potential of Cannabidiol for the Treatment of Viral Hepatitis. Pharmacogn. Res. 2017, 9, 116–118. [Google Scholar] [CrossRef] [Green Version]
- Maor, Y.; Yu, J.; Kuzontkoski, P.M.; Dezube, B.J.; Zhang, X.; Groopman, J.E. Cannabidiol Inhibits Growth and Induces Programmed Cell Death in Kaposi Sarcoma–Associated Herpesvirus-Infected Endothelium. Genes Cancer 2012, 3, 512–520. [Google Scholar] [CrossRef] [Green Version]
- Morales, P.; Muller, C.; Jagerovic, N.; Reggio, P.H. Targeting CB2 and TRPV1: Computational Approaches for the Identification of Dual Modulators. Front. Mol. Biosci. 2022, 9, 841190. [Google Scholar] [CrossRef]
- DeMarino, C.; Barclay, R.A.; Pleet, M.L.; Pinto, D.O.; Branscome, H.; Paul, S.; Lepene, B.; El-Hage, N.; Kashanchi, F. Purification of High Yield Extracellular Vesicle Preparations Away from Virus. J. Vis. Exp. JoVE 2019, 151, e59876. [Google Scholar] [CrossRef]







| EV Subpopulation | CBD | HU308 | |
|---|---|---|---|
| EV Concentration | 2K | ## | ### |
| 10K | ### | - | |
| 100K | *** | *** | |
| 167K(S) | - | ** | |
| 167K(L) | ### | - | |
| Median Size | 2K | - | - |
| 10K | - | - | |
| 100K | - | * | |
| 167K(S) | - | ## | |
| 167K(L) | ** | *** | |
| Mean Size | 2K | - | - |
| 10K | - | * | |
| 100K | - | * | |
| 167K(S) | - | ## | |
| 167K(L) | - | ** | |
| Peak Size | 2K | - | - |
| 10K | - | - | |
| 100K | - | - | |
| 167K(S) | - | - | |
| 167K(L) | * | ** | |
| TAR | 2K | - | ## |
| 10K | - | ## | |
| 100K | *** | ** | |
| 167K(S) | ** | * | |
| 167K(L) | # | - | |
| env | 2K | - | - |
| 10K | - | - | |
| 100K | ** | ** | |
| 167K(S) | - | - | |
| 167K(L) | # | * | |
| TNF-α | 2K | * | *** |
| 10K | # | - | |
| 100K | - | - | |
| 167K(S) | - | - | |
| 167K(L) | # | - | |
| IL-1β | 2K | - | *** |
| 10K | # | ## | |
| 100K | - | - | |
| 167K(S) | # | - | |
| 167K(L) | # | - | |
| Primary Macrophage+ cART (low) | TAR | *** | *** |
| TAR-gag | *** | - | |
| env | *** | *** | |
| Primary Macrophage | TAR | *** | *** |
| TAR-gag | ** | ** | |
| env | *** | *** | |
| Neurosphere | TAR | *** | *** |
| TAR-gag | ** | *** | |
| env | * | *** | |
| IL-1β | ### | * | |
| TNF-α | ### | * | |
| Mouse | HU308 + cART (low) | TAR | ** |
| TAR-gag | * | ||
| env | ** | ||
| HU308 | TAR | ** | |
| TAR-gag | * | ||
| env | * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Williams, A.; Khatkar, P.; Branscome, H.; Kim, Y.; Erickson, J.; Jenabian, M.-A.; Costiniuk, C.T.; Kashanchi, F. The Use of CBD and Its Synthetic Analog HU308 in HIV-1-Infected Myeloid Cells. Pharmaceuticals 2023, 16, 1147. https://doi.org/10.3390/ph16081147
Williams A, Khatkar P, Branscome H, Kim Y, Erickson J, Jenabian M-A, Costiniuk CT, Kashanchi F. The Use of CBD and Its Synthetic Analog HU308 in HIV-1-Infected Myeloid Cells. Pharmaceuticals. 2023; 16(8):1147. https://doi.org/10.3390/ph16081147
Chicago/Turabian StyleWilliams, Anastasia, Pooja Khatkar, Heather Branscome, Yuriy Kim, James Erickson, Mohammad-Ali Jenabian, Cecilia T. Costiniuk, and Fatah Kashanchi. 2023. "The Use of CBD and Its Synthetic Analog HU308 in HIV-1-Infected Myeloid Cells" Pharmaceuticals 16, no. 8: 1147. https://doi.org/10.3390/ph16081147