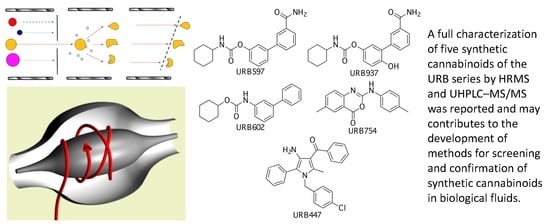
Characterization of URB Series Synthetic Cannabinoids by HRMS and UHPLC–MS/MS
Abstract
:1. Introduction
2. Results and Discussion
2.1. URB597
2.2. URB937
2.3. URB602
2.4. URB754
2.5. URB447
3. Materials and Methods
3.1. Standard and Solvents
3.2. Chemicals
3.3. UHPLC-MS/MS
3.4. HRMS Compound Characterization
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, P.; Mahato, D.K.; Kamle, M.; Borah, R.; Sharma, B.; Pandhi, S.; Tripathi, V.; Yadav, H.S.; Devi, S.; Patil, U.; et al. Pharmacological properties, therapeutic potential, and legal status of Cannabis sativa L.: An overview. Phytother. Res. 2021, 35, 6010–6029. [Google Scholar] [CrossRef] [PubMed]
- Pagano, C.; Navarra, G.; Coppola, L.; Avilia, G.; Bifulco, M.; Laezza, C. Cannabinoids: Therapeutic use in clinical practice. Int. J. Mol. Sci. 2022, 23, 3344. [Google Scholar] [CrossRef] [PubMed]
- Gülck, T.; Møller, B.L. Phytocannabinoids: Origins and biosynthesis. Trends Plant Sci. 2020, 25, 985–1004. [Google Scholar] [CrossRef] [PubMed]
- European Monitoring Centre for Drugs and Drug Addiction. European Drug Reports. Trends and Developments. 2022. Available online: https://www.emcdda.europa.eu/system/files/publications/14644/TDAT22001ENN.pdf (accessed on 21 December 2022).
- Devane, W.A.; Dysarz, F.A.; Johnson, M.R.; Melvin, L.S.; Howlett, A.C. Determination and characterization of a cannabinoid receptor in rat brain. Mol. Pharmacol. 1988, 34, 605–613. [Google Scholar] [PubMed]
- Matsuda, L.A.; Lolait, S.J.; Brownstein, M.J.; Young, A.C.; Bonner, T.I. Structure of a cannabinoid receptor and functional ex-pression of the cloned cDNA. Nature 1990, 346, 561–564. [Google Scholar] [CrossRef]
- Munro, S.; Thomas, K.L.; Abu-Shaar, M. Molecular characterization of a peripheral receptor for cannabinoids. Nature 1993, 365, 61–65. [Google Scholar] [CrossRef]
- Lutz, B. Neurobiology of cannabinoid receptor signaling. Dialogues Clin. Neurosci. 2020, 22, 207–222. [Google Scholar] [CrossRef]
- Shahbazi, F.; Grandi, V.; Banerjee, A.; Trant, J.F. Cannabinoids and cannabinoid receptors: The story so far. iScience 2020, 23, 101301. [Google Scholar] [CrossRef]
- Ren, S.; Wang, Z.; Zhang, Y.; Chen, N. Potential application of endocannabinoid system agents in neuropsychiatric and neuro-degenerative diseases—Focusing on FAAH/MAGL inhibitors. Acta Pharmacol. Sin. 2020, 41, 1263–1271. [Google Scholar] [CrossRef]
- Van Sickle, M.D.; Duncan, M.; Kingsley, P.J.; Mouihate, A.; Urbani, P.; Mackie, K.; Stella, N.; Makriyannis, A.; Piomelli, D.; Davison, J.S.; et al. Identification and functional characterization of brainstem cannabinoid CB2 receptors. Science 2005, 310, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Liu, Z.; Li, X.; Wang, J.; Li, L. Cannabinoid receptors in myocardial injury: A brother born to rival. Int. J. Mol. Sci. 2021, 22, 6886. [Google Scholar] [CrossRef] [PubMed]
- Devane, W.A.; Hanus, L.; Breuer, A.; Pertwee, R.G.; Stevenson, L.A.; Griffin, G.; Gibson, D.; Mandelbaum, A.; Etinger, A.; Mech-oulam, R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science 1992, 258, 1946–1949. [Google Scholar] [CrossRef]
- Mechoulam, R.; Ben-Shabat, S.; Hanus, L.; Ligumsky, M.; Kaminski, N.E.; Schatz, A.R.; Gopher, A.; Almog, S.; Martin, B.R.; Compton, D.R. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem. Pharmacol. 1995, 50, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, T.; Kondo, S.; Sukagawa, A.; Nakane, S.; Shinoda, A.; Itoh, K.; Yamashita, A.; Waku, K. 2-Arachidonoylglycerol: A possible endogenous cannabinoid receptor ligand in brain. Biochem. Biophys. Res. Commun. 1995, 215, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Piomelli, D. The molecular logic of endocannabinoid signalling. Nat. Rev. Neurosci. 2003, 4, 873–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Augustin, S.M.; Lovinger, D.M. Functional Relevance of endocannabinoid-dependent synaptic plasticity in the central nervous system. ACS Chem. Neurosci. 2018, 9, 2146–2161. [Google Scholar] [CrossRef]
- Lowe, H.; Toyang, N.; Steele, B.; Bryant, J.; Ngwa, W. The endocannabinoid system: A potential target for the treatment of various diseases. Int. J. Mol. Sci. 2021, 22, 9472. [Google Scholar] [CrossRef]
- Piomelli, D.; Mabou Tagne, A. Endocannabinoid-based therapies. Ann. Rev. Pharmacol. Toxicol. 2022, 62, 17.1–17.25. [Google Scholar] [CrossRef]
- Gaoni, Y.; Mechoulam, R. Isolation, structure, and partial synthesis of an active constituent of hashish. J. Am. Chem. Soc. 1964, 86, 1646–1647. [Google Scholar] [CrossRef]
- Peng, J.; Fan, M.; An, C.; Ni, F.; Huang, W.; Luo, J. A narrative review of molecular mechanism and therapeutic effect of cannabidiol (CBD). Basic Clin. Pharmacol. Toxicol. 2022, 130, 439–456. [Google Scholar] [CrossRef]
- Manera, C.; Bertini, S. Cannabinoid-Based Medicines and Multiple Sclerosis. Adv. Exp. Med. Biol. 2021, 1264, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Jazz Pharmaceutics Website. Available online: https://www.jazzpharma.com/medicines/our-medicines/ (accessed on 21 December 2022).
- Pertwee, R.G. Targeting the endocannabinoid system with cannabinoid receptor agonists: Pharmacological strategies and therapeutic possibilities. Phil. Trans. R. Soc. B 2012, 367, 3353–3363. [Google Scholar] [CrossRef] [Green Version]
- De Luca, M.A.; Fattore, L. Therapeutic use of synthetic cannabinoids: Still an open issue? Clin. Ther. 2018, 40, 1457–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, D.; Peigneur, S.; Hendrickx, L.A.; Tytgat, J. Targeting cannabinoid receptors: Current status and prospects of natural prod-ucts. Int. J. Mol. Sci. 2020, 21, 5064. [Google Scholar] [CrossRef] [PubMed]
- Estrada, J.A.; Contreras, I. Endocannabinoid receptors in the CNS: Potential drug targets for the prevention and treatment of neurologic and psychiatric disorders. Curr. Neuropharmacol. 2020, 18, 769–787. [Google Scholar] [CrossRef]
- Wiley, J.L.; Compton, D.R.; Dai, D.; Lainton, J.A.H.; Phillips, M.; Huffman, J.W.; Martin, B.R. Structure-activity relationships of indole- and pyrrole-derived cannabinoids. J. Pharmacol. Exp. Ther. 1998, 285, 995–1004. [Google Scholar]
- Auwärter, V.; Dresen, S.; Weinmann, W.; Müller, M.; Pütz, M.; Ferreirós, N. ‘Spice’ and other herbal blends: Harmless incense or cannabinoid designer drugs? J. Mass Spectrom. 2009, 44, 832–837. [Google Scholar] [CrossRef]
- Atwood, B.K.; Huffman, J.; Straiker, A.; Mackie, K. JWH018, a common constituent of ‘Spice’ herbal blends, is a potent and efficacious cannabinoid CB receptor agonist. Br. J. Pharmacol. 2010, 160, 585–593. [Google Scholar] [CrossRef] [Green Version]
- UNODC Early Warning Advisory Toxicology Highlights. Current NPS Threats, Volume II, January 2020. Available online: https://www.unodc.org/documents/scientific/Current_NPS_Threats_Volume_II_Web.pdf (accessed on 21 December 2022).
- Shafi, A.; Berry, A.J.; Sumnall, H.; Wood, D.M.; Tracy, D.K. New psychoactive substances: A review and updates. Ther. Adv. Psychopharmacol. 2020, 10, 2045125320967197. [Google Scholar] [CrossRef] [PubMed]
- Giorgetti, A.; Pascali, J.P.; Fai, P.; Pelletti, G.; Gabbin, A.; Franchetti, G.; Cecchetto, G.; Viel, G. Molecular mechanisms of action of novel psychoactive substances (NPS). A new threat for young drug users with forensic-toxicological implications. Life 2021, 11, 440. [Google Scholar] [CrossRef]
- UNODC Early Warning Advisory Toxicology Highlights. Current NPS Threats, Volume IV. November 2021. Available online: https://www.unodc.org/documents/scientific/NPS_threats-IV.pdf (accessed on 21 December 2022).
- Chung, E.Y.; Cha, H.J.; Min, H.K.; Yun, J. Pharmacology and adverse effects of new psychoactive substances: Synthetic cannabinoid receptor agonists. Arch. Pharm. Res. 2021, 44, 402–413. [Google Scholar] [CrossRef] [PubMed]
- UNODC Early Warning Advisory on New Psychoactive Substances. What Are NPS? December 2021. Available online: https://www.unodc.org/LSS/Page/NPS (accessed on 21 December 2022).
- European Monitoring Centre for Drugs and Drug Addiction. Synthetic Cannabinoids in Europe—A Review. 2021. Available online: https://www.emcdda.europa.eu/system/files/publications/14035/Synthetic-cannabinoids-in-Europe-EMCDDA-technical-report.pdf (accessed on 21 December 2022).
- Markham, J.; Sparkes, E.; Boyd, R.; Chen, S.; Manning, J.J.; Finlay, D.; Lai, F.; McGregor, E.; Maloney, C.J.; Gerona, R.R.; et al. Defining steric requirements at CB1 and CB2 cannabinoid receptors using synthetic cannabinoid receptor agonists 5F-AB-PINACA, 5F-ADB-PINACA, PX-1, PX-2, NNL-1, and their analogues. ACS Chem. Neurosci. 2022, 13, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Alves, V.L.; Gonçalves, J.L.; Aguiar, J.; Teixeira, H.M.; Câmara, J.S. The synthetic cannabinoids phenomenon: From structure to toxicological properties. A review. Crit. Rev. Toxicol. 2020, 50, 359–382. [Google Scholar] [CrossRef] [PubMed]
- Kathuria, S.; Gaetani, S.; Fegley, D.; Valiño, F.; Duranti, A.; Tontini, A.; Mor, M.; Tarzia, G.; La Rana, G.; Calignano, A.; et al. Modulation of anxiety through blockade of anandamide hydrolysis. Nat. Med. 2003, 9, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Clapper, J.R.; Moreno-Sanz, G.; Russo, R.; Guijarro, A.; Vacondio, F.; Duranti, A.; Tontini, A.; Sanchini, S.; Sciolino, N.R.; Spradley, J.M.; et al. Anandamide suppresses pain initiation through a peripheral endocannabinoid mechanism. Nat. Neurosci. 2010, 13, 1265–1270. [Google Scholar] [CrossRef] [Green Version]
- Hohmann, A.G.; Suplita, R.L.; Bolton, N.M.; Neely, M.H.; Fegley, D.; Mangieri, R.; Krey, J.F.; Walker, J.M.; Holmes, P.V.; Crystal, J.D.; et al. An endocannabinoid mechanism for stress-induced analgesia. Nature 2005, 435, 1108–1112. [Google Scholar] [CrossRef] [Green Version]
- Makara, J.K.; Mor, M.; Fegley, D.; Szabó, S.I.; Kathuria, S.; Astarita, G.; Duranti, A.; Tontini, A.; Tarzia, G.; Rivara, S.; et al. Selective inhibition of 2-AG hydrolysis enhances endocannabinoid signaling in hippocampus. Nat. Neurosci. 2005, 8, 1139–1141. [Google Scholar] [CrossRef]
- Tarzia, G.; Antonietti, F.; Duranti, A.; Tontini, A.; Mor, M.; Rivara, S.; Traldi, P.; Astarita, G.; King, A.; Clapper, J.R.; et al. Identification of a bioactive impurity in a commercial sample of 6-methyl-2-p-tolylaminobenzo[d][1,3]oxazin-4-one (URB754). Ann. Chim. 2007, 97, 887–894. [Google Scholar] [CrossRef] [Green Version]
- LoVerme, J.; Duranti, A.; Tontini, A.; Spadoni, G.; Mor, M.; Rivara, S.; Stella, N.; Xu, C.; Tarzia, G.; Piomelli, D. Synthesis and characterization of a peripherally restricted CB1 cannabinoid antagonist, URB447, that reduces feeding and body-weight gain in mice. Bioorg. Med. Chem. Lett. 2009, 19, 639–643. [Google Scholar] [CrossRef] [Green Version]
- Benedicto, A.; Arteta, B.; Duranti, A.; Alonso-Alconada, D. The synthetic cannabinoid URB447 exerts antitumor and antimetastatic effect in melanoma and colon cancer. Pharmaceuticals 2022, 15, 1166. [Google Scholar] [CrossRef]
- Nakajima, J.; Takahashi, M.; Seto, T.; Kanai, C.; Suzuki, J.; Yoshida, M.; Uemura, N.; Hamano, T. Analysis of azepane isomers of AM-2233 and AM-1220, and detection of an inhibitor of fatty acid amide hydrolase [3′-(aminocarbonyl)(1,1′-biphenyl)-3-yl]-cyclohexylcarbamate (URB597) obtained as designer drugs in the Tokyo area. Forensic Toxicol. 2013, 31, 76–85. [Google Scholar] [CrossRef]
- Shanks, K.G.; Behonick, G.S.; Dahn, T.; Terrell, A. Identification of novel third-generation synthetic cannabinoids in products by ultra-performance liquid chromatography and time-of-flight mass spectrometry. J. Anal. Toxicol. 2013, 37, 517–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: http://www.emcdda.europa.eu/attachements.cfm/att_229598_EN_TDAN14001ENN.pdf (accessed on 21 December 2022).
- Uchiyama, N.; Kawamura, M.; Kikura-Hanajiri, R.; Goda, Y. URB-754: A new class of designer drug and 12 synthetic cannabinoids detected in illegal products. Forensic Sci. Int. 2013, 227, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Tarzia, G.; Duranti, A.; Tontini, A.; Piersanti, G.; Mor, M.; Rivara, S.; Plazzi, P.V.; Park, C.; Kathuria, S.; Piomelli, D. Design, synthesis, and structure–activity relationships of alkylcarbamic acid aryl esters, a new class of fatty acid amide hydrolase inhibitors. J. Med. Chem. 2003, 46, 2352–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mor, M.; Rivara, S.; Lodola, A.; Plazzi, P.V.; Tarzia, G.; Duranti, A.; Tontini, A.; Piersanti, G.; Kathuria, S.; Piomelli, D. Cyclohexylcarbamic Acid 3′- or 4′-substituted biphenyl-3-yl esters as fatty acid amide hydrolase inhibitors: Synthesis, quantitative structure-activity relationships, and molecular modelling studies. J. Med. Chem. 2004, 47, 4998–5008. [Google Scholar] [CrossRef]
- Available online: https://namsdl.org/wp-content/uploads/Summary-of-Synthetic-Drugs-Bills-and-Proposed-Regulations.pdf (accessed on 16 January 2023).




















| Compound (Elementary Formula) | RT | [M + H]+ (m/z) | Product Ions (m/z) | QTT/QLT Ion Ratio (%) | DP (V) | EP (V) | CE (V) | CXP (V) |
|---|---|---|---|---|---|---|---|---|
| JWH-018 (C24H23NO) (internal standard) | 5.22 | 342 | 155 | QTT | 110 | 10 | 35 | 11 |
| 127 | 60 | 110 | 10 | 70 | 6 | |||
| 145 | 33 | 110 | 10 | 59 | 11 | |||
| URB597 (C20H22N2O3) | 3.16 | 339 | 214 | QTT | 78 | 10 | 18 | 11 |
| 197 | 62 | 78 | 10 | 31 | 17 | |||
| 171 | 30 | 78 | 10 | 39 | 12 | |||
| URB937 | 2.62 | 355 | 338 | 6 | 80 | 10 | 16 | 8 |
| (C20H22N2O4) | 230 | QTT | 80 | 10 | 22 | 5 | ||
| 213 | 90 | 80 | 10 | 32 | 11 | |||
| URB602 (C19H21NO2) | 4.84 | 296 | 214 | QTT | 45 | 10 | 14 | 10 |
| 196 | 50 | 45 | 10 | 29 | 15 | |||
| 170 | 67 | 45 | 10 | 28 | 15 | |||
| URB754 (C16H14N2O2) | 2.36 | 267 | 249 | 63 | 70 | 10 | 34 | 18 |
| 160 | QTT | 70 | 10 | 32 | 11 | |||
| 106 | 6 | 70 | 10 | 40 | 7 | |||
| URB447 (C25H21ClN2O) | 3.43 | 401 | 296 | 45 | 90 | 10 | 34 | 7 |
| 276 | 25 | 90 | 10 | 31 | 6 | |||
| 105 | QTT | 90 | 10 | 38 | 5 | |||
| 403 | 298 | 14 | 90 | 10 | 32 | 7 |
| Compound | Elementary Formula (MH+) | Observed m/z | Theoretical m/z | Δppm |
|---|---|---|---|---|
| URB597 | C20H23N2O3+ | 339.1703 | 339.1703 | – |
| C13H12NO2+ | 214.0861 | 214.0862 | −0.5 | |
| C13H9O2+ | 197.0596 | 197.0597 | −0.5 | |
| C12H11O+ | 171.0801 | 171.0804 | −1.7 | |
| URB937 | C20H20NO4Na+ | 377.1474 | 377.1471 | 0.8 |
| C20H20NO4+ | 338.1384 | 338.1387 | −0.9 | |
| C13H12NO3+ | 230.0809 | 230.0812 | −1.3 | |
| C13H9O3+ | 213.0544 | 213.0546 | −0.9 | |
| URB602 | C19H22NO2+ | 296.1646 | 296.1650 | −1.3 |
| URB754 | C16H15N2O2+ | 267.1128 | 267.1128 | – |
| C16H13N2O+ | 249.1023 | 249.1022 | 0.4 | |
| C9H6NO2+ | 160.0391 | 160.0393 | −1.2 | |
| C7H8N+ | 106.0648 | 106.0651 | −2.8 | |
| URB447 | C25H22ClN2O+ | 401.1413 | 401.1415 | −0.5 |
| C18H17N2Cl+ | 296.1073 | 296.1074 | 0.3 | |
| C18H16N2O+ | 276.1256 | 276.1257 | −0.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agostini, M.; Favretto, D.; Renzoni, C.; Vogliardi, S.; Duranti, A. Characterization of URB Series Synthetic Cannabinoids by HRMS and UHPLC–MS/MS. Pharmaceuticals 2023, 16, 201. https://doi.org/10.3390/ph16020201
Agostini M, Favretto D, Renzoni C, Vogliardi S, Duranti A. Characterization of URB Series Synthetic Cannabinoids by HRMS and UHPLC–MS/MS. Pharmaceuticals. 2023; 16(2):201. https://doi.org/10.3390/ph16020201
Chicago/Turabian StyleAgostini, Marco, Donata Favretto, Caterina Renzoni, Susanna Vogliardi, and Andrea Duranti. 2023. "Characterization of URB Series Synthetic Cannabinoids by HRMS and UHPLC–MS/MS" Pharmaceuticals 16, no. 2: 201. https://doi.org/10.3390/ph16020201