Momordica cochinchinensis (Gấc) Seed Extracts Induce Apoptosis and Necrosis in Melanoma Cells
Abstract
:1. Introduction
2. Results
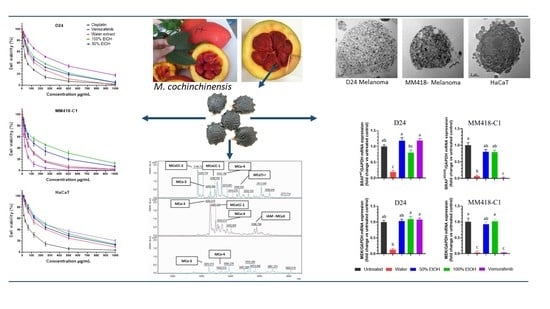
2.1. Viability and Toxicity on Cancer and Control Cells
2.2. Morphological Cellular Changes
2.3. Upregulation of TNFR1 Expression by M. cochinchinensis Seed Extracts Induced Melanoma Cell Death through the Suppression of the Nuclear Translocation of NF-kB
2.4. M. cochinchinensis Seed Extracts Supressed Expression of MAPK Genes in the Melanoma Cells
2.5. Proteins and Peptides in M. cochinchinensis Seed Extracts
2.6. Determination of Potential Oncogene Proteins from M. cochinchinensis Seed
2.7. Correlations between Cytotoxicity, Extraction Solvent, Gene Expression and Phytochemicals
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of Extracts
4.3. Cell Culture Initial Growth
4.4. Cytotoxicity Assays
4.5. Morphological Changes of Cells Using Transmission Electron Micrography (TEM)
4.6. Gene Expression of TNFR1, NF-kB, BRAF, MEK1 and Nrf2
4.7. Protein and Peptide Profile Analysis
4.8. SDS-PAGE Electrophoresis
4.9. Putative Peptides Detected Using MALDI-TOF-MS
4.10. Bioinformatic Analysis of Potential Oncogene Proteins
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ottaviano, M.; Giunta, E.; Tortora, M.; Curvietto, M.; Attademo, L.; Bosso, D.; Cardalesi, C.; Rosanova, M.; de Placido, P.; Pietroluongo, E.; et al. BRAF Gene and Melanoma: Back to the Future. Int. J. Mol. Sci. 2021, 22, 3474. [Google Scholar] [CrossRef] [PubMed]
- Malissen, N.; Grob, J.-J. Metastatic Melanoma: Recent Therapeutic Progress and Future Perspectives. Drugs 2018, 78, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Berthenet, K.; Ferrer, C.C.; Fanfone, D.; Popgeorgiev, N.; Neves, D.; Bertolino, P.; Gibert, B.; Hernandez-Vargas, H.; Ichim, G. Failed Apoptosis Enhances Melanoma Cancer Cell Aggressiveness. Cell Rep. 2020, 31, 107731. [Google Scholar] [CrossRef]
- Chan, X.Y.; Singh, A.; Osman, N.; Piva, T.J. Role Played by Signalling Pathways in Overcoming BRAF Inhibitor Resistance in Melanoma. Int. J. Mol. Sci. 2017, 18, 1527. [Google Scholar] [CrossRef]
- Johnson, D.B.; Sosman, J.A. BRAFV600E-mutant melanoma presenting with cardiac involvement. J. Natl. Compr. Cancer Netw. 2014, 12, 611–615. [Google Scholar] [CrossRef] [Green Version]
- Tas, F.; Erturk, K. BRAF V600E mutation as a prognostic factor in cutaneous melanoma patients. Dermatol. Ther. 2020, 33, e13270. [Google Scholar] [CrossRef]
- Schmidlin, C.J.; Tian, W.; Dodson, M.; Chapman, E.; Zhang, D.D. FAM129B-dependent activation of NRF2 promotes an invasive phenotype in BRAF mutant melanoma cells. Mol. Carcinog. 2021, 60, 331–341. [Google Scholar] [CrossRef]
- Kreß, J.; Jessen, C.; Marquardt, A.; Hufnagel, A.; Meierjohann, S. NRF2 Enables EGFR Signaling in Melanoma Cells. Int. J. Mol. Sci. 2021, 22, 3803. [Google Scholar] [CrossRef]
- Lehraiki, A.; Cerezo, M.; Rouaud, F.; Abbe, P.; Allegra, M.; Kluza, J.; Marchetti, P.; Imbert, V.; Cheli, Y.; Bertolotto, C.; et al. Increased CD271 expression by the NF-kB pathway promotes melanoma cell survival and drives acquired resistance to BRAF inhibitor vemurafenib. Cell Discov. 2015, 1, 15030. [Google Scholar] [CrossRef] [Green Version]
- Eberle, J. Countering TRAIL Resistance in Melanoma. Cancers 2019, 11, 656. [Google Scholar] [CrossRef] [Green Version]
- Seca, A.M.L.; Pinto, D.C.G.A. Plant Secondary Metabolites as Anticancer Agents: Successes in Clinical Trials and Therapeutic Application. Int. J. Mol. Sci. 2018, 19, 263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wimalasiri, D.; Piva, T.; Huynh, T. Diversity in Nutrition and Bioactivity of Momordica cochinchinensis. Int. J. Adv. Sci. Eng. Inf. Technol. 2016, 6, 378. [Google Scholar] [CrossRef]
- Huynh, T.; Nguyen, M.H. Bioactive compounds from Gac (Momordica cochinchinensis Lour. Spreng). In Bioactive Compounds in Underutilized Fruits and Nuts; Murthy, H., Bapat., V., Eds.; Springer: Cham, Switzerland, 2019; Volume 11–12, pp. 591–604. [Google Scholar] [CrossRef]
- Cheung, S.C.; Li, N.H. Chinese Medicinal Herbs of Hong Kong; Commercial Press: Shanghai, China, 1997. [Google Scholar] [CrossRef]
- Wong, K.L.; Wong, R.N.S.; Zhang, L.; Liu, W.K.; Ng, T.B.; Shaw, P.C.; Kwok, P.C.L.; Lai, Y.M.; Zhang, Z.J.; Zhang, Y.; et al. Bioactive proteins and peptides isolated from Chinese medicines with pharmaceutical potential. Chin. Med. 2014, 9, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tien, P.G.; Kayama, F.; Konishi, F.; Tamemoto, H.; Kasono, K.; Hung, N.T.K.; Kuroki, M.; Ishikawa, S.-E.; Van, C.N.; Kawakami, M. Inhibition of tumor growth and angiogenesis by water extract of Gac fruit (Momordica cochinchinensis Spreng). Int. J. Oncol. 2005, 26, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhang, Y.; Liu, Y.; Yang, X.O.; Zhan, Y. Momordica cochinchinensis Spreng. seed extract suppresses breast cancer growth by inducing cell cycle arrest and apoptosis. Mol. Med. Rep. 2015, 12, 6300–6310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, L.Y.; Liu, H.R.; Shen, Y.; Yu, Y.Q.; Tao, X. Cochinchina momordica seed extract induces G2/M arrest and apoptosis in human breast cancer MDA-MB-231 cells by modulating the PI3K/Akt pathway. APJCP 2012, 13, 3483–3488. [Google Scholar]
- Liu, H.-R.; Meng, L.-Y.; Lin, Z.-Y.; Shen, Y.; Yu, Y.-Q.; Zhu, Y.-Z. Cochinchina momordica Seed Extract Induces Apoptosis and Cell Cycle Arrest in Human Gastric Cancer Cells Via PARP and p53 Signal Pathways. Nutr. Cancer 2012, 64, 1070–1077. [Google Scholar] [CrossRef]
- Shen, Y.; Meng, L.; Sun, H.; Zhu, Y.; Liu, H. Cochinchina momordica Seed Suppresses Proliferation and Metastasis in Human Lung Cancer Cells by Regulating Multiple Molecular Targets. Am. J. Chin. Med. 2015, 43, 149–166. [Google Scholar] [CrossRef]
- Ma, M.; Zhao, L.-M.; Yang, X.-X.; Shan, Y.-N.; Cui, W.-X.; Chen, L.; Shan, B.-E. p-Hydroxylcinnamaldehyde induces the differentiation of oesophageal carcinoma cells via the cAMP-RhoA-MAPK signalling pathway. Sci. Rep. 2016, 6, 31315. [Google Scholar] [CrossRef]
- Zhao, L.-M.; Sun, G.-G.; Han, L.-N.; Liu, L.-H.; Ren, F.-Z.; Li, L.; Ma, M.; Shan, B.-E. P-Hydroxycinnamaldehyde Induces B16-F1 Melanoma Cell Differentiation via the RhoA-MAPK Signaling Pathway. Cell. Physiol. Biochem. 2016, 38, 2247–2260. [Google Scholar] [CrossRef] [PubMed]
- Le, A.V.; Huynh, T.T.; Parks, S.E.; Nguyen, M.H.; Roach, P.D. Bioactive Composition, Antioxidant Activity, and Anticancer Potential of Freeze-Dried Extracts from Defatted Gac (Momordica cochinchinensis Spreng) Seeds. Medicines 2018, 5, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, L.Y.; Wang, C.K.L.; Major, J.M.; Greenwood, K.P.; Lewis, R.J.; Craik, D.J.; Daly, N.L. Isolation and Characterization of Peptides from Momordica cochinchinensis Seeds. J. Nat. Prod. 2009, 72, 1453–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolognesi, A.; Barbieri, L.; Abbondanza, A.; Falasca, A.I.; Carnicelli, D.; Battelli, M.G.; Stirpe, F. Purification and properties of new ribosome-inactivating proteins with RNA N-glycosidase activity. Biochim. Biophys. Acta Gene Struct. Expr. 1990, 1087, 293–302. [Google Scholar] [CrossRef]
- Chuethong, J.; Oda, K.; Sakurai, H.; Saiki, I.; Leelamanit, W. Cochinin B, a Novel Ribosome-Inactivating Protein from the Seeds of Momordica cochinchinensis. Biol. Pharm. Bull. 2007, 30, 428–432. [Google Scholar] [CrossRef] [Green Version]
- Dean, L. Vemurafenib Therapy and BRAF and NRAS Genotype. In Medical Genetics Summaries; Pratt, V.M., Scott, S.A., Pirmohamed, M., Esquivel, B., Kattman, B.L., Malheiro, A.J., Eds.; National Center for Biotechnology Information: Bethesda, MD, USA, 2017. Available online: https://www.ncbi.nlm.nih.gov/books/NBK447416/ (accessed on 24 December 2021).
- D’Arcy, M.S. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 2019, 43, 582–592. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Montfort, A.; Colacios, C.; Levade, T.; Andrieu-Abadie, N.; Meyer, N.; Ségui, B. The TNF Paradox in Cancer Progression and Immunotherapy. Front. Immunol. 2019, 10, 1818. [Google Scholar] [CrossRef] [Green Version]
- Jaco, I.; Annibaldi, A.; Lalaoui, N.; Wilson, R.; Tenev, T.; Laurien, L.; Kim, C.; Jamal, K.; John, S.W.; Liccardi, G.; et al. MK2 Phosphorylates RIPK1 to Prevent TNF-Induced Cell Death. Mol. Cell 2017, 66, 698–710.e5. [Google Scholar] [CrossRef]
- Josephs, S.F.; Ichim, T.E.; Prince, S.M.; Kesari, S.; Marincola, F.M.; Escobedo, A.R.; Jafri, A. Unleashing endogenous TNF-alpha as a cancer immunotherapeutic. J. Transl. Med. 2018, 16, 242–250. [Google Scholar] [CrossRef] [Green Version]
- Jessen, C.; Kreß, J.K.C.; Baluapuri, A.; Hufnagel, A.; Schmitz, W.; Kneitz, S.; Roth, S.; Marquardt, A.; Appenzeller, S.; Ade, C.P.; et al. The transcription factor NRF2 enhances melanoma malignancy by blocking differentiation and inducing COX2 expression. Oncogene 2020, 39, 6841–6855. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.Y.; He, W.; Tan, N.; Zeng, G.; Craik, D.J.; Daly, N.L. A new family of cystine knot peptides from the seeds of Momordica cochinchinensis. Peptides 2013, 39, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Yu, Q.; Gong, Y.; Chen, W.; Tong, Y.; Wang, Y.; Xu, H.; Shi, Y. Yes-Associated Protein (YAP) Promotes Tumorigenesis in Melanoma Cells Through Stimulation of Low-Density Lipoprotein Receptor-Related Protein 1 (LRP1). Sci. Rep. 2017, 7, 15528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salama, Y.; Lin, S.-Y.; Dhahri, D.; Hattori, K.; Heissig, B. The fibrinolytic factor tPA drives LRP1-mediated melanoma growth and metastasis. FASEB J. 2018, 33, 3465–3480. [Google Scholar] [CrossRef]
- Zhou, H.; Ekmekcioglu, S.; Marks, J.W.; Mohamedali, K.A.; Asrani, K.; Phillips, K.K.; Brown, S.A.; Cheng, E.; Weiss, M.B.; Hittelman, W.N.; et al. The TWEAK Receptor Fn14 Is a Therapeutic Target in Melanoma: Immunotoxins Targeting Fn14 Receptor for Malignant Melanoma Treatment. J. Investig. Dermatol. 2013, 133, 1052–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lassen, U.N.; Meulendijks, D.; Siu, L.L.; Karanikas, V.; Mau-Sorensen, M.; Schellens, J.H.; Jonker, D.J.; Hansen, A.R.; Simcox, M.E.; Schostack, K.J.; et al. A Phase I Monotherapy Study of RG7212, a First-in-Class Monoclonal Antibody Targeting TWEAK Signaling in Patients with Advanced Cancers. Clin. Cancer Res. 2015, 21, 258–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, A.S.K.; Tchounwou, P.B. Cisplatin-based chemotherapy of human cancers. J. Cancer Sci. Ther. 2020, 11, 97. [Google Scholar]
- Yu, J.S.; Roh, H.-S.; Lee, S.; Jung, K.; Baek, K.-H.; Kim, K.H. Antiproliferative effect of Momordica cochinchinensis seeds on human lung cancer cells and isolation of the major constituents. Rev. Bras. Farm. 2017, 27, 329–333. [Google Scholar] [CrossRef]
- Castellano, M.; Pollock, P.; Walters, M.K.; E Sparrow, L.; Down, L.M.; Gabrielli, B.; Parsons, P.; Hayward, N.K. CDKN2A/p16 is inactivated in most melanoma cell lines. Cancer Res. 1997, 57, 4868–4875. [Google Scholar]
- Peltzer, N.; Darding, M.; Walczak, H. Holding RIPK1 on the Ubiquitin Leash in TNFR1 Signaling. Trends Cell Biol. 2016, 26, 445–461. [Google Scholar] [CrossRef]
- Patiño, J.A.G.; Ivanov, V.N.; Lacy, E.; Elkon, K.B.; Marino, M.W.; Nikolić-Žugić, J. TNF-α Is the Critical Mediator of the Cyclic AMP-Induced Apoptosis of CD8+4+Double-Positive Thymocytes. J. Immunol. 2000, 164, 1689–1694. [Google Scholar] [CrossRef] [PubMed]
- Kahroba, H.; Shirmohamadi, M.; Hejazi, M.S.; Samadi, N. The Role of Nrf2 signaling in cancer stem cells: From stemness and self-renewal to tumorigenesis and chemoresistance. Life Sci. 2019, 239, 116986. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, C.; Qin, L.; Xu, J.; Li, X.; Wang, W.; Kong, L.; Zhou, T.; Li, X. The prognostic value of NRF2 in solid tumor patients: A meta-analysis. Oncotarget 2017, 9, 1257–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, N.J.; Hinner, M.J. Getting Across the Cell Membrane: An Overview for Small Molecules, Peptides, and Proteins. In Site-Specific Protein Labeling: Methods and Protocols; Methods in Molecular Biology Book Series; Gautier, A., Hinner, M.J., Eds.; Springer: New York, NY, USA, 2015; Volume 1266, pp. 29–53. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Xiao, S.; Xia, Y. Tumor Necrosis Factor Receptor Mediates Fibroblast Growth Factor-Inducible 14 Signaling. Cell. Physiol. Biochem. 2017, 43, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Wimalasiri, D.; Dekiwadia, C.; Fong, S.Y.; Piva, T.J.; Huynh, T. Anticancer activity of Momordica cochinchinensis (red gac) aril and the impact of varietal diversity. BMC Complement. Med. Ther. 2020, 20, 365. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Zhou, F.; Hearne, Z.; Li, C.-J. Water—The greenest solvent overall. Curr. Opin. Green Sustain. Chem. 2019, 18, 118–123. [Google Scholar] [CrossRef]








| Proteins (kDa) | Protein Concentration (ng/mg) | ||
|---|---|---|---|
| Water | 50% EtOH | 100% EtOH | |
| 20.0 | 18.9 ± 1.98 a | 12.7 ± 1.32 b | 3.4 ± 0.97 c |
| 30.0 | 7.9 ± 1.03 a | 4.8 ± 0.88 b | 1.2 ± 0.63 c |
| 35.1 | 8.7 ± 0.93 a | 4.5 ± 0.76 b | 1.8 ± 0.64 c |
| 36.7 | 6.5 ± 1.09 a | 3.8 ± 0.79 b | 1.3 ± 0.82 c |
| 55.1 | 1.7 ± 0.54 a | 0.00 b | 0.00 b |
| 64.3 | 2.1 ± 0.35 a | 0.00 b | 0.00 b |
| Primer | Forward Sequence (5′-3′) | Reverse Sequence (5′-3′) | Amplification Efficiency (E) (%) |
|---|---|---|---|
| TNFR1 | GGGCACCTTTACGGCTTCC | GGTTCTCCTTACAGCCACACA | 99 |
| NF-kB | ATAGAAGAGCAGCGTGGGGACT | GGATGACGTAAAGGGATAGGGC | 98 |
| Nrf2 | AGTGGATCTGCCAACTACTC | CATCTACAAACGGGAATGTCTG | 99 |
| BRAFWT | GGCAGAGTGCCTCAAAAAGAA | AACCAGCCCGATTCAAGGA | 99 |
| BRAFV600E | CCGACCAGCAGATGAAGATCAT | TCAACATTTTCACTGCCACATCAC | 99 |
| MEK1 | CAGAAGAAGCTGGAGGAGCTAG | CCATCGCTGTAGAACGCACCAT | 97 |
| GADPH | TCCACCACCCTGTTGCTGTA | ACCACAGTCCATGCCATCAC | 99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, D.; Holien, J.; Dekiwadia, C.; Thrimawithana, T.; Piva, T.; Huynh, T. Momordica cochinchinensis (Gấc) Seed Extracts Induce Apoptosis and Necrosis in Melanoma Cells. Pharmaceuticals 2023, 16, 100. https://doi.org/10.3390/ph16010100
Nguyen D, Holien J, Dekiwadia C, Thrimawithana T, Piva T, Huynh T. Momordica cochinchinensis (Gấc) Seed Extracts Induce Apoptosis and Necrosis in Melanoma Cells. Pharmaceuticals. 2023; 16(1):100. https://doi.org/10.3390/ph16010100
Chicago/Turabian StyleNguyen, Dao, Jessica Holien, Chaitali Dekiwadia, Thilini Thrimawithana, Terrence Piva, and Tien Huynh. 2023. "Momordica cochinchinensis (Gấc) Seed Extracts Induce Apoptosis and Necrosis in Melanoma Cells" Pharmaceuticals 16, no. 1: 100. https://doi.org/10.3390/ph16010100