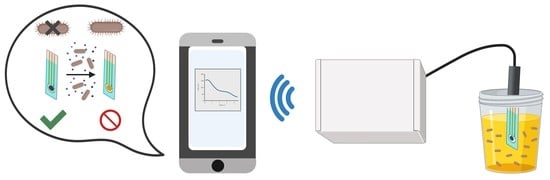
Portable Prussian Blue-Based Sensor for Bacterial Detection in Urine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Preparation and Characterization of PrBl-Modified SPEs
2.3. Wireless Sensing
2.4. Bacteria Cultivation
2.5. Flow Cytometry
2.6. Scanning Electron Microscopy
3. Results and Discussion
3.1. Electrodeposition and Characterization of PrBl-Modified SPE
3.2. Wireless Sensing Principle Using PrBl-Modified SPEs
3.3. Wireless Sensing in AUM and Human Urine
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmed, A.; Rushworth, J.V.; Hirst, N.A.; Millner, P.A. Biosensors for Whole-Cell Bacterial Detection. Clin. Microbiol. Rev. 2014, 27, 631–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, M.L.; Gaido, L. Laboratory Diagnosis of Urinary Tract Infections in Adult Patients. Med. Microbiol. 2004, 38, 1150–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabe, M.; Bjerklund-Johansen, T.E.; Botto, H.; Çek, M.; Naber, K.G.; Tenke, P.; Wagenlehner, F. Guidelines on Urological Infections. Eur. Assoc. Urol. 2015, 182, 237–257. [Google Scholar]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Ipe, D.S.; Horton, E.; Ulett, G.C. The Basics of Bacteriuria: Strategies of Microbes for Persistence in Urine. Front. Cell. Infect. Microbiol. 2016, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- McLellan, L.K.; Hunstad, D.A. Urinary Tract Infection: Pathogenesis and Outlook. Trends Mol. Med. 2016, 22, 946–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smelov, V.; Naber, K.; Johansen, T.E.B. Improved Classification of Urinary Tract Infection: Future Considerations. Eur. Urol. Suppl. 2016, 15, 71–80. [Google Scholar] [CrossRef]
- Lam, C.-W.; Law, C.-Y.; To, K.K.-W.; Cheung, S.K.-K.; Lee, K.-C.; Sze, K.-H.; Leung, K.-F.; Yuen, K.-Y. NMR-based metabolomic urinalysis: A rapid screening test for urinary tract infection. Clin. Chim. Acta 2014, 436, 217–223. [Google Scholar] [CrossRef]
- Karlsen, H.; Dong, T. Biomarkers of urinary tract infections: State of the art, and promising applications for rapid strip-based chemical sensors. Anal. Methods 2015, 7, 7961–7975. [Google Scholar] [CrossRef]
- Nicolle, L.E.; SHEA Long-Term–Care Committee. Urinary tract infections in long-term–care facilities. Infect. Control. Hosp. Epidemiol. 2001, 22, 167–175. [Google Scholar] [CrossRef]
- Chin, C.D.; Linder, V.; Sia, S.K. Lab-on-a-chip devices for global health: Past studies and future opportunities. Lab Chip 2007, 7, 41–57. [Google Scholar] [CrossRef]
- Schmiemann, G.; Gágyor, I.; Hummers-Pradier, E.; Bleidorn, J. Resistance profiles of urinary tract infections in general practice—An observational study. BMC Urol. 2012, 12, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Yu, L.; Liu, Y.; Xu, H.; Ma, L.; Tian, P.; Zhu, J.; Wang, F.; Yi, K.; Xiao, H.; et al. Space-time-regulated imaging analyzer for smart coagulation diagnosis. Cell Rep. Med. 2022, 3, 100765. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Y.; Xu, H.; Ma, L.; Wang, Y.; Yu, L.; Wang, F.; Zhu, J.; Hu, X.; Yi, K.; et al. Touchable cell biophysics property recognition platforms enable multifunctional blood smart health care. Microsyst. Nanoeng. 2021, 7, 103. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-M.; Won, D.D.; Lee, B.J.; Escobedo, D.; Esteva, A.; Aalipour, A.; Ge, T.J.; Kim, J.H.; Suh, S.; Choi, E.H.; et al. A mountable toilet system for personalized health monitoring via the analysis of excreta. Nat. Biomed. Eng. 2020, 4, 624–635. [Google Scholar] [CrossRef]
- Beduk, T.; Beduk, D.; Hasan, M.R.; Celik, E.G.; Kosel, J.; Narang, J.; Salama, K.N.; Timur, S. Smartphone-Based Multiplexed Biosensing Tools for Health Monitoring. Biosensors 2022, 12, 583. [Google Scholar] [CrossRef]
- Karyakin, A.A. Prussian blue and its analogues: Electrochemistry and analytical applications. Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal. 2001, 13, 813–819. [Google Scholar] [CrossRef]
- Karyakin, A.A. Advances of Prussian blue and its analogues in (bio)sensors. Curr. Opin. Electrochem. 2017, 5, 92–98. [Google Scholar] [CrossRef]
- Pearce, J. Studies of any toxicological effects of Prussian blue compounds in mammals—A review. Food Chem. Toxicol. 1994, 32, 577–582. [Google Scholar] [CrossRef]
- Zhao, J.; Cai, X.; Gao, W.; Zhang, L.; Zou, D.; Zheng, Y.; Li, Z.; Chen, H. Prussian Blue Nanozyme with Multienzyme Activity Reduces Colitis in Mice. ACS Appl. Mater. Interfaces 2018, 10, 26108–26117. [Google Scholar] [CrossRef]
- Zhang, W.; Hu, S.; Yin, J.-J.; He, W.; Lu, W.; Ma, M.; Gu, N.; Zhang, Y. Prussian Blue Nanoparticles as Multienzyme Mimetics and Reactive Oxygen Species Scavengers. J. Am. Chem. Soc. 2016, 138, 5860–5865. [Google Scholar] [CrossRef] [PubMed]
- De la Paz, E.; Barfidokht, A.; Rios, S.; Brown, C.; Chao, E.; Wang, J. Extended Noninvasive Glucose Monitoring in the Interstitial Fluid Using an Epidermal Biosensing Patch. Anal. Chem. 2021, 93, 12767–12775. [Google Scholar] [CrossRef] [PubMed]
- Sempionatto, J.R.; Moon, J.-M.; Wang, J. Touch-Based Fingertip Blood-Free Reliable Glucose Monitoring: Personalized Data Processing for Predicting Blood Glucose Concentrations. ACS Sensors 2021, 6, 1875–1883. [Google Scholar] [CrossRef] [PubMed]
- Pilas, J.; Selmer, T.; Keusgen, M.; Schöning, M.J. Screen-Printed Carbon Electrodes Modified with Graphene Oxide for the Design of a Reagent-Free NAD+-Dependent Biosensor Array. Anal. Chem. 2019, 91, 15293–15299. [Google Scholar] [CrossRef]
- Morris, K.; Zhao, H.; John, R. Ferricyanide-Mediated Microbial Reactions for Environmental Monitoring. Aust. J. Chem. 2005, 58, 237–245. [Google Scholar] [CrossRef]
- Jahn, M.K.; Haderlein, S.B.; Meckenstock, R.U. Reduction of Prussian Blue by the two iron-reducing microorganisms Geobacter metallireducens and Shewanella alga. Environ. Microbiol. 2006, 8, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Vilanova, A.; Alonso, Y.; Dietvorst, J.; Pérez-Montero, M.; Rodríguez-Rodríguez, R.; Ivanova, K.; Tzanov, T.; Vigués, N.; Mas, J.; Guirado, G.; et al. Sonochemical coating of Prussian Blue for the production of smart bacterial-sensing hospital textiles. Ultrason. Sonochemistry 2021, 70, 105317. [Google Scholar] [CrossRef]
- Ferrer-Vilanova, A.; Alonso, Y.; Ezenarro, J.J.; Santiago, S.; Muñoz-Berbel, X.; Guirado, G. Electrochromogenic Detection of Live Bacteria Using Soluble and Insoluble Prussian Blue. ACS Omega 2021, 6, 30989–30997. [Google Scholar] [CrossRef]
- Zelada-Guillén, G.A.; Bhosale, S.V.; Riu, J.; Rius, F.X. Real-Time Potentiometric Detection of Bacteria in Complex Samples. Anal. Chem. 2010, 82, 9254–9260. [Google Scholar] [CrossRef]
- Poma, N.; Vivaldi, F.; Bonini, A.; Salvo, P.; Kirchhain, A.; Melai, B.; Bottai, D.; Tavanti, A.; Di Francesco, F. A graphenic and potentiometric sensor for monitoring the growth of bacterial biofilms. Sensors Actuators B Chem. 2020, 323, 128662. [Google Scholar] [CrossRef]
- Brooks, T.; Keevil, C. A simple artificial urine for the growth of urinary pathogens. Lett. Appl. Microbiol. 1997, 24, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M. Short Protocols in Molecular Biology, 5th ed.; Sonns, J.W., Ed.; Wiley: Hoboken, NJ, USA, 2002; Volume 1, pp. 1–6. [Google Scholar]
- Moshaver, B.; de Boer, F.; van Egmond-Kreileman, H.; Kramer, E.; Stegeman, C.; Groeneveld, P. Fast and accurate prediction of positive and negative urine cultures by flow cytometry. BMC Infect. Dis. 2016, 16, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baggio, B.F.; Vicente, C.; Pelegrini, S.; Cid, C.C.P.; Brandt, I.S.; Tumelero, M.A.; Pasa, A.A. Morphology and Structure of Electrodeposited Prussian Blue and Prussian White Thin Films. Materials 2019, 12, 1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koncki, R. Chemical Sensors and Biosensors Based on Prussian Blues. Crit. Rev. Anal. Chem. 2002, 32, 79–96. [Google Scholar] [CrossRef]
- Kulesza, P.J.; Doblhofer, K. The membrane properties of Prussian Blue films on electrodes. J. Electroanal. Chem. Interfacial Electrochem. 1989, 274, 95–105. [Google Scholar] [CrossRef]
- Beal, J.; Farny, N.G.; Haddock-Angelli, T.; Selvarajah, V.; Baldwin, G.S.; Buckley-Taylor, R.; Gershater, M.; Kiga, D.; Marken, J.; Sanchania, V. Robust estimation of bacterial cell count from optical density. Commun. Biol. 2020, 3, 512. [Google Scholar] [CrossRef]
- Robador, A.; LaRowe, D.E.; Finkel, S.E.; Amend, J.P.; Nealson, K.H. Changes in Microbial Energy Metabolism Measured by Nanocalorimetry during Growth Phase Transitions. Front. Microbiol. 2018, 9, 109. [Google Scholar] [CrossRef]
- Makinoshima, H.; Nishimura, A.; Ishihama, A. Fractionation of Escherichia coli cell populations at different stages during growth transition to stationary phase. Mol. Microbiol. 2002, 43, 269–279. [Google Scholar] [CrossRef] [Green Version]
- Unden, G.; Dünnwald, P. The Aerobic and Anaerobic Respiratory Chain of Escherichia coli and Salmonella enterica: Enzymes and Energetics. EcoSal Plus 2008, 3, 6. [Google Scholar] [CrossRef]
- Qiao, Y.; Li, C.M.; Bao, S.-J.; Lu, Z.; Hong, Y. Direct electrochemistry and electrocatalytic mechanism of evolved Escherichia coli cells in microbial fuel cells. Chem. Commun. 2008, 44, 1290–1292. [Google Scholar] [CrossRef]
- Wang, L.; Fan, D.; Chen, W.; Terentjev, E. Bacterial growth, detachment and cell size control on polyethylene terephthalate surfaces. Sci. Rep. 2015, 5, 15159. [Google Scholar] [CrossRef] [PubMed]
- Tuson, H.H.; Weibel, D.B. Bacteria–surface interactions. Soft Matter 2013, 9, 4368–4380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubitschek, H.E. Buoyant Density Variation during the Cell Cycle in Microorganisms. CRC Crit. Rev. Microbiol. 1987, 14, 73–97. [Google Scholar] [CrossRef]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli Physiology in Luria-Bertani Broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, B.; Selomulya, C.; Zheng, G.; Zhao, D. New faces of porous Prussian blue: Interfacial assembly of integrated hetero-structures for sensing applications. Chem. Soc. Rev. 2015, 44, 7997–8018. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.; Mediati, D.G.; Duggin, I.G.; Harry, E.J.; Bottomley, A.L. Metabolic Adaptations of Uropathogenic E. coli in the Urinary Tract. Front. Cell. Infect. Microbiol. 2017, 7, 241. [Google Scholar] [CrossRef] [PubMed]




| Compound | Quantity (g) |
|---|---|
| Peptone L37 | 1 |
| Yeast extract | 0.005 |
| Lactic acid | 0.1 |
| Citric acid | 0.4 |
| Sodium bicarbonate | 2.1 |
| Urea | 10 |
| Uric acid | 0.07 |
| Creatinine | 0.8 |
| Calcium chloride | 0.37 |
| Sodium chloride | 5.2 |
| Iron II sulphate | 0.0012 |
| Magnesium sulphate | 0.49 |
| Sodium sulphate | 3.2 |
| Potassium dihydrogen phosphate | 0.95 |
| Potassium hydrogen phosphate | 1.2 |
| Ammonium chloride | 1.3 |
| Distilled water | to 1 L |
| Incubation Time (h) | OCP Unmodified SPE in 200 µL Bacteria (mV) | OCP PrBl SPE in 200 µL Bacteria (mV) | OCP PrBl SPE No Bacteria (mV) | OCP PrBl SPE in 2 µL Bacteria (mV) | Bacteria Concentration in 2 µL (Flow Cytometry) (Cells/mL) |
|---|---|---|---|---|---|
| 0 | 28 | 141 | 141 | 125 | 7 × 103 |
| 1 | 88 | 80 | 101 | 109 | |
| 2 | 72 | 51 | 92 | 92 | 1.5 × 104 |
| 3 | 79 | 24 | 94 | 91 | |
| 4 | 82 | −1 | 92 | 80 | 2.4 × 105 |
| 5 | 57 | −31 | 93 | 78 | |
| 6 | 40 | −59 | 94 | 26 | 4.2 × 106 |
| 7 | 57 | −86 | 90 | −6 | |
| 23 | 61 | −168 | 99 | −41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Psotta, C.; Chaturvedi, V.; Gonzalez-Martinez, J.F.; Sotres, J.; Falk, M. Portable Prussian Blue-Based Sensor for Bacterial Detection in Urine. Sensors 2023, 23, 388. https://doi.org/10.3390/s23010388
Psotta C, Chaturvedi V, Gonzalez-Martinez JF, Sotres J, Falk M. Portable Prussian Blue-Based Sensor for Bacterial Detection in Urine. Sensors. 2023; 23(1):388. https://doi.org/10.3390/s23010388
Chicago/Turabian StylePsotta, Carolin, Vivek Chaturvedi, Juan F. Gonzalez-Martinez, Javier Sotres, and Magnus Falk. 2023. "Portable Prussian Blue-Based Sensor for Bacterial Detection in Urine" Sensors 23, no. 1: 388. https://doi.org/10.3390/s23010388