Surface Topography, Microbial Adhesion, and Immune Responses in Silicone Mammary Implant-Associated Capsular Fibrosis
Abstract
:1. Introduction
2. Results
2.1. Impact of Surface Roughness on Antimicrobial Response and Wound Proteome Adhesion on SMI Surface
2.2. Quantification and Data Integrity of Intraindividual Comparative Microbiome Profiling in Wound Bed Fluid, Capsular Tissue, and SMI-Adhesive Microbiome
2.3. Microbial Dynamics in the Peri-Implant Environment: A Comprehensive Analysis of Skin Microbiome Transfer and Biofilm Formation during Implant-Based Breast Reconstruction
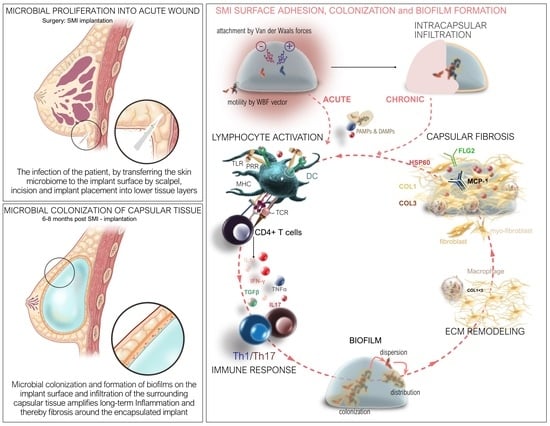
2.4. Staphylococcal Transmission in Surgery: Skin Microbiota Transfer, Biofilm Formation, and Chronic Inflammation Leading to Implant Encapsulation
2.5. In Vitro Evaluation of Silicone SMI Surface Topography Impact on Staphylococci Adhesion, Growth, Colonization, and Biofilm Formation
2.6. Immunoreactivity of Staphylococci Biofilms on Silicone Implant Surfaces In Vitro
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Study Design
4.3. Biological Sample Collection
4.4. The Mass Spectrometry Proteomics Data Source
4.4.1. Identification, Characterization, and Quantification of Differential Common and Topography-Exclusive Wound Bed Proteome
4.4.2. Identification and Characterization of Common and Topography-Exclusive Adsorbed Wound Bed Proteome on SMI Surface
4.5. Microbiome Profiling by Next-Generation DNA Sequencing
4.5.1. Sample Preparation
4.5.2. Control Measures for Microbiome Analysis: Normalization of Wound Bed Fluid, SMI Surface, and Capsular Tissue Data to Multiple Negative Controls
4.5.3. Statistical Analyses
4.6. Validation of Sequencing Data: Cultivation and MALDI TOF Identification of Skin Swab Samples from Surgical Incision Sites and Proximal Regions
4.7. Microbial Culture and Biofilm Formation on SMI Surfaces In Vitro
4.7.1. SMI Material and Sterilization
4.7.2. Biofilm Formation and Bacteria
4.7.3. Biofilm Dissolution and Quantification
4.7.4. Scanning Electron Microscopy
4.8. Bacterial Biofilm Gene Expression and PBMC Inflammatory Gene Expression Using Real-Time qPCR
4.8.1. Cell Isolation and In Vitro Culture with Biofilm-Associated SMI Surfaces
4.8.2. Reverse Transcription Real-Time Quantitative PCR (RT-qPCR)
4.9. Statistics
4.10. Ethics Statement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ghazal, I.D.; Eleweke, C.; Aladesanwa, F.O.; Onabajo, B.O. Post-mastectomy breast reconstruction: An overview of the state of the art, challenges, and prospects. Int. Surg. J. 2023, 10, 348–354. [Google Scholar] [CrossRef]
- Perry, D.; Frame, J. The history and development of breast implants. Ind. Mark. Manag. 2020, 102, 478–482. [Google Scholar] [CrossRef]
- Saini, M.; Singh, Y.; Arora, P.; Arora, V.; Jain, K. Implant biomaterials: A comprehensive review. World J. Clin. Cases WJCC 2015, 3, 52. [Google Scholar] [CrossRef]
- Mempin, M.; Hu, H.; Chowdhury, D.; Deva, A.; Vickery, K. The A, B and C’s of silicone breast implants: Anaplastic large cell lymphoma, biofilm and capsular contracture. Materials 2018, 11, 2393. [Google Scholar] [CrossRef] [PubMed]
- Bizjak, M.; Selmi, C.; Praprotnik, S.; Bruck, O.; Perricone, C.; Ehrenfeld, M.; Shoenfeld, Y. Silicone implants and lymphoma: The role of inflammation. J. Autoimmun. 2015, 65, 64–73. [Google Scholar] [CrossRef]
- Wick, G.; Backovic, A.; Rabensteiner, E.; Plank, N.; Schwentner, C.; Sgonc, R. The immunology of fibrosis: Innate and adaptive responses. Trends Immunol. 2010, 31, 110–119. [Google Scholar] [CrossRef]
- Wick, G.; Grundtman, C.; Mayerl, C.; Wimpissinger, T.F.; Feichtinger, J.; Zelger, B.; Sgonc, R.; Wolfram, D. The immunology of fibrosis. Annu. Rev. Immunol. 2013, 31, 107–135. [Google Scholar] [CrossRef] [PubMed]
- Bachour, Y.; Verweij, S.P.; Gibbs, S.; Ket, J.C.; Ritt, M.J.; Niessen, F.B.; Mullender, M.G. The aetiopathogenesis of capsular contracture: A systematic review of the literature. J. Plast. Reconstr. Aesthetic Surg. 2018, 71, 307–317. [Google Scholar] [CrossRef]
- Prantl, L.; Schreml, S.; Fichtner-Feigl, S.; Pöppl, N.; Eisenmann-Klein, M.; Schwarze, H.; Füchtmeier, B. Clinical and morphological conditions in capsular contracture formed around silicone breast implants. Plast. Reconstr. Surg. 2007, 120, 275–284. [Google Scholar] [CrossRef]
- Safran, T.; Nepon, H.; Chu, C.K.; Winocour, S.; Murphy, A.M.; Davison, P.G.; Dionisopolos, T.; Vorstenbosch, J. Healing, Inflammation, and Fibrosis: Current Concepts in Capsular Contracture: Pathophysiology, Prevention, and Management. Semin. Plast. Surg. 2021, 35, 189. [Google Scholar]
- Siggelkow, W.; Faridi, A.; Spiritus, K.; Klinge, U.; Rath, W.; Klosterhalfen, B. Histological analysis of silicone breast implant capsules and correlation with capsular contracture. Biomaterials 2003, 24, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Siggelkow, W.; Gescher, D.; Siggelkow, A.; Klee, D.; Malik, E.; Rath, W.; Faridi, A. In vitro analysis of modified surfaces of silicone breast implants. Int. J. Artif. Organs 2004, 27, 1100–1108. [Google Scholar] [CrossRef] [PubMed]
- Handel, N.; Jensen, J.A.; Black, Q.; Waisman, J.R.; Silverstein, M.J. The fate of breast implants: A critical analysis of complications and outcomes. Plast. Reconstr. Surg. 1995, 96, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A. Cellular and molecular mechanisms of fibrosis. J. Pathol. 2008, 214, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Wang, T.; Tian, L.; Song, H.; Gao, M. Roxatidine inhibits fibrosis by inhibiting NF κB and MAPK signaling in macrophages sensing breast implant surface materials. Mol. Med. Rep. 2020, 21, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Kuehlmann, B.A.; Bonham, C.A.; Gurtner, G.C. Abstract 114. Targeting Wnt Signaling to Reduce Capsular Fibrosis. Plast. Reconstr. Surg. Glob. Open 2019, 7 (Suppl. S4), 80. [Google Scholar] [CrossRef]
- Kuo, Y.-L.; Jou, I.-M.; Jeng, S.-F.; Chu, C.-H.; Huang, J.-S.; Hsu, T.-I.; Chang, L.-R.; Huang, P.-W.; Chen, J.-A.; Chou, T.-M. Hypoxia-induced epithelial-mesenchymal transition and fibrosis for the development of breast capsular contracture. Sci. Rep. 2019, 9, 10269. [Google Scholar] [CrossRef]
- Meng, X.-M.; Nikolic-Paterson, D.J.; Lan, H.Y. TGF-β: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef]
- Araújo-Gomes, N.; Romero-Gavilán, F.; Sánchez-Pérez, A.M.; Gurruchaga, M.; Azkargorta, M.; Elortza, F.; Martinez-Ibañez, M.; Iloro, I.; Suay, J.; Goni, I. Characterization of serum proteins attached to distinct sol–gel hybrid surfaces. J. Biomed. Mater. Res. Part B Appl. Biomater. 2018, 106, 1477–1485. [Google Scholar] [CrossRef]
- Backovic, A.; Wolfram, D.; Del-Frari, B.; Piza, H.; Huber, L.A.; Wick, G. Simultaneous analysis of multiple serum proteins adhering to the surface of medical grade polydimethylsiloxane elastomers. J. Immunol. Methods 2007, 328, 118–127. [Google Scholar] [CrossRef]
- Schoberleitner, I.; Faserl, K.; Sarg, B.; Egle, D.; Brunner, C.; Wolfram, D. Quantitative Proteomic Characterization of Foreign Body Response towards Silicone Breast Implants Identifies Chronological Disease-Relevant Biomarker Dynamics. Biomolecules 2023, 13, 305. [Google Scholar] [CrossRef]
- Backovic, A.; Huang, H.-L.; Del Frari, B.; Piza, H.; Huber, L.A.; Wick, G. Identification and dynamics of proteins adhering to the surface of medical silicones in vivo and in vitro. J. Proteome Res. 2006, 6, 376–381. [Google Scholar] [CrossRef]
- Doloff, J.C.; Veiseh, O.; de Mezerville, R.; Sforza, M.; Perry, T.A.; Haupt, J.; Jamiel, M.; Chambers, C.; Nash, A.; Aghlara-Fotovat, S.; et al. The surface topography of silicone breast implants mediates the foreign body response in mice, rabbits and humans. Nat. Biomed. Eng. 2021, 5, 1115–1130. [Google Scholar] [CrossRef] [PubMed]
- ISO 14607:2018; Non-Active Surgical Implants—Mammary Implants—Particular Requirements. Available online: https://www.iso.org/standard/63973.html (accessed on 1 March 2024).
- Jalalabadi, F.; Doval, A.F.; Neese, V.B.; Andrews, E.B.; Spiegel, A.J. Breast Implant Utilization Trends in USA versus Europe and the Impact of BIA-ALCL Publications. Plast. Reconstr. Surg.-Glob. Open 2021, 9, e3449. [Google Scholar] [CrossRef]
- Keech, J.A.; Creech, B.J. Anaplastic t-cell lymphoma in proximity to a saline-filled breast implant. Plast. Reconstr. Surg. 1997, 100, 554–555. [Google Scholar] [CrossRef] [PubMed]
- Tevis, S.E.; Hunt, K.K.; Miranda, R.N.; Lange, C.; Pinnix, C.C.; Iyer, S.; Butler, C.E.; Clemens, M.W. Breast Implant-associated Anaplastic Large Cell Lymphoma: A Prospective Series of 52 Patients. Ann. Surg. 2022, 275, E245–E249. [Google Scholar] [CrossRef] [PubMed]
- Barnsley, G.P.; Sigurdson, L.J.; Barnsley, S.E. Textured surface breast implants in the prevention of capsular contracture among breast augmentation patients: A meta-analysis of randomized controlled trials. Plast. Reconstr. Surg. 2006, 117, 2182–2190. [Google Scholar] [CrossRef]
- Cappellano, G.; Ploner, C.; Lobenwein, S.; Sopper, S.; Hoertnagl, P.; Mayerl, C.; Wick, N.; Pierer, G.; Wick, G.; Wolfram, D. Immunophenotypic characterization of human T cells after in vitro exposure to different silicone breast implant surfaces. PLoS ONE 2018, 13, e0192108. [Google Scholar] [CrossRef]
- Schoberleitner, I.; Faserl, K.; Tripp, C.H.; Pechriggl, E.J.; Sigl, S.; Brunner, A.; Hermann-Kleiter, N.; Baier, L.; Steinkellner, T.; Sarg, B.; et al. Silicone implant surface microtopography modulates inflammation and tissue repair in capsular fibrosis. Front. Immunol. 2024, 15. [Google Scholar] [CrossRef]
- Schoberleitner, I.; Augustin, A.; Egle, D.; Brunner, C.; Amort, B.; Zelger, B.; Brunner, A.; Wolfram, D. Is It All about Surface Topography? An Intra-Individual Clinical Outcome Analysis of Two Different Implant Surfaces in Breast Reconstruction. J. Clin. Med. 2023, 12, 1315. [Google Scholar] [CrossRef]
- Mohapatra, S. Sterilization and Disinfection. Essent. Neuroanesthesia 2017, 929. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7158362/ (accessed on 1 March 2024).
- Belay, E.D.; Schonberger, L.B.; Brown, P.; Priola, S.A.; Chesebro, B.; Will, R.G.; Asher, D.M. Disinfection and Sterilization of Prion-Contaminated Medical Instruments. Infect. Control Hosp. Epidemiol. 2010, 31, 1304–1306. [Google Scholar] [CrossRef]
- Rutala, W.A.; Weber, D.J. Guideline for Disinfection and Sterilization of Prion-Contaminated Medical Instruments. Infect. Control Hosp. Epidemiol. 2010, 31, 107–117. [Google Scholar] [CrossRef]
- Dapunt, U.; Prior, B.; Kretzer, J.P.; Giese, T.; Zhao, Y. Bacterial Biofilm Components Induce an Enhanced Inflammatory Response Against Metal Wear Particles. Ther. Clin. Risk Manag. 2020, ume 16, 1203–1212. [Google Scholar] [CrossRef]
- Marcinkiewicz, J.; Strus, M.; Pasich, E. Antibiotic resistance: A “dark side” of biofilm-associated chronic infections. Pol. Arch. Med. Wewn. 2013, 123, 309–313. [Google Scholar] [CrossRef]
- Carvajal, J.; Carvajal, M.; Hernández, G. Back to Basics: Could the Preoperative Skin Antiseptic Agent Help Prevent Biofilm-Related Capsular Contracture? Aesthetic Surg. J. 2019, 39, 848–859. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Rieger, U.M.; Mesina, J.; Kalbermatten, D.F.; Haug, M.; Frey, H.P.; Pico, R.; Frei, R.; Pierer, G.; Lüscher, N.J.; Trampuz, A. Bacterial biofilms and capsular contracture in patients with breast implants. Br. J. Surg. 2013, 100, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Ajdic, D.; Zoghbi, Y.; Gerth, D.; Panthaki, Z.J.; Thaller, S. The relationship of bacterial biofilms and capsular contracture in breast implants. Aesthetic Surg. J. 2016, 36, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.-J.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef]
- Deva, A.K.; Adams, W.P., Jr.; Vickery, K. The role of bacterial biofilms in device-associated infection. Plast. Reconstr. Surg. 2013, 132, 1319–1328. [Google Scholar] [CrossRef]
- del Pozo, J.L.; Auba, C. Role of biofilms in breast implant associated infections and capsular contracture. Adv. Exp. Med. Biol. 2015, 831, 53–67. [Google Scholar] [PubMed]
- Pajkos, A.; Deva, A.K.; Vickery, K.; Cope, C.; Chang, L.; Cossart, Y.E. Detection of subclinical infection in significant breast implant capsules. Plast. Reconstr. Surg. 2003, 111, 1605–1611. [Google Scholar] [CrossRef]
- Adams, W.P.; Culbertson, E.J.; Deva, A.K.; Magnusson, M.R.; Layt, C.; Jewell, M.L.; Mallucci, P.; Hedén, P. Macrotextured breast implants with defined steps to minimize bacterial contamination around the device: Experience in 42,000 implants. Plast. Reconstr. Surg. 2017, 140, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Gofstein-Hayuth, D.; Fliss, E.; Barnea, Y.; Legarda, C.; Bracha, G.; Lerner, A.; Lellouche, J.; Carmeli, Y.; Shani, N.; Arad, E. Comparing the efficacy of antimicrobial pocket-irrigation protocols in an in vivo breast implant infection model. J. Plast. Reconstr. Aesthetic Surg. 2023, 85, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Ibarburu, B.; Díaz-Navarro, M.; Ibarra, G.; Rivera, A.; Hafian, R.; Irigoyen, Ã; Carrillo, R.; Pérez-Cano, R.; Muñoz, P.; García-Ruano, Á.; et al. Efficacy of Povidone Iodine Against Microbial Biofilms in Breast Implants With Different Textures: Results From an in vitro Study. Front. Microbiol. 2022, 13, 868347. [Google Scholar] [CrossRef]
- Cometta, S.; Bock, N.; Suresh, S.; Dargaville, T.R.; Hutmacher, D.W. Antibacterial Albumin-Tannic Acid Coatings for Scaffold-Guided Breast Reconstruction. Front. Bioeng. Biotechnol. 2021, 9, 638577. [Google Scholar] [CrossRef]
- Lam, M.; Migonney, V.; Falentin-Daudre, C. Review of silicone surface modification techniques and coatings for antibacterial/antimicrobial applications to improve breast implant surfaces. Acta Biomater. 2020, 121, 68–88. [Google Scholar] [CrossRef]
- Panczel, G.; Munhoz, A.M. A Simple and Low-cost Method of Sleeve to Insert Silicone Gel Breast Implants. Plast. Reconstr. Surg.-Glob. Open 2019, 7, e2389. [Google Scholar] [CrossRef]
- Burkhardt, B.R.; Fried, M.; Schnur, P.L.; Tofield, J.J. Capsules, infection, and intraluminal antibiotics. Plast Reconstr. Surg. 1981, 68, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Shah, Z.; Lehman, J.A.; Tan, J. Does infection play a role in breast capsular contracture? Plast Reconstr. Surg. 1981, 68, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Moon, D.J.; Deva, A.K. Adverse Events Associated with Breast Implants: The Role of Bacterial Infection and Biofilm. Clin. Plast. Surg. 2021, 48, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Virden, C.P.; Dobke, M.K.; Stein, P.; Parsons, C.L.; Frank, D.H. Subclinical Infection of the Silicone Breast Implant Surface as a Possible Cause of Capsular Contracture. Aesthetic Plast. Surg. 2020, 44, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Deva, A.; Chang, L. Bacterial biofilms: A cause for accelerated capsular contracture? Aesthetic Surg. J. 1999, 19, 130–133. [Google Scholar] [CrossRef]
- Fey, P.D.; Olson, M.E. Current concepts in biofilm formation of Staphylococcus epidermidis. Futur. Microbiol. 2010, 5, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Moris, V.; Lam, M.; Amoureux, L.; Magallon, A.; Guilloteau, A.; Maldiney, T.; Zwetyenga, N.; Falentin-Daudre, C.; Neuwirth, C. What is the best technic to dislodge Staphylococcus epidermidis biofilm on medical implants? BMC Microbiol. 2022, 22, 192. [Google Scholar] [CrossRef] [PubMed]
- Rezende-Pereira, G.; Albuquerque, J.P.; Souza, M.C.; A Nogueira, B.; Silva, M.G.; Hirata, R.; Mattos-Guaraldi, A.L.; Duarte, R.S.; Neves, F.P.G. Biofilm Formation on Breast Implant Surfaces by Major Gram-Positive Bacterial Pathogens. Aesthetic Surg. J. 2020, 41, 1144–1151. [Google Scholar] [CrossRef]
- Mu, M.; Liu, S.; DeFlorio, W.; Hao, L.; Wang, X.; Salazar, K.S.; Taylor, M.; Castillo, A.; Cisneros-Zevallos, L.; Oh, J.K.; et al. Influence of Surface Roughness, Nanostructure, and Wetting on Bacterial Adhesion. Langmuir 2023, 39, 5426–5439. [Google Scholar] [CrossRef]
- Brauman, D. Rough Textured Silicone Implants, Bacterial Biofilms, and Capsular Contracture. Plast. Reconstr. Surg. 2019, 144, 322e–323e. [Google Scholar] [CrossRef]
- James, G.A.; Boegli, L.; Hancock, J.; Bowersock, L.; Parker, A.; Kinney, B.M. Bacterial Adhesion and Biofilm Formation on Textured Breast Implant Shell Materials. Aesthetic Plast. Surg. 2018, 43, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Encinas, N.; Yang, C.-Y.; Geyer, F.; Kaltbeitzel, A.; Baumli, P.; Reinholz, J.; Mailänder, V.; Butt, H.-J.; Vollmer, D. Submicrometer-Sized Roughness Suppresses Bacteria Adhesion. ACS Appl. Mater. Interfaces 2020, 12, 21192–21200. [Google Scholar] [CrossRef] [PubMed]
- Yoda, I.; Koseki, H.; Tomita, M.; Shida, T.; Horiuchi, H.; Sakoda, H.; Osaki, M. Effect of surface roughness of biomaterials on Staphylococcus epidermidis adhesion. BMC Microbiol. 2014, 14, 234. [Google Scholar] [CrossRef] [PubMed]
- Alessandri-Bonetti, M.; Jeong, T.; Vaienti, L.; De La Cruz, C.; Gimbel, M.L.; Nguyen, V.T.; Egro, F.M. The Role of Microorganisms in the Development of Breast Implant-Associated Anaplastic Large Cell Lymphoma. Pathogens 2023, 12, 313. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef]
- Seykora, J.; Dentchev, T.; Margolis, D.J. Filaggrin-2 barrier protein inversely varies with skin inflammation. Exp. Dermatol. 2015, 24, 720–722. [Google Scholar] [CrossRef] [PubMed]
- Hansmann, B.; Schröder, J.-M.; Gerstel, U. Skin-Derived C-Terminal Filaggrin-2 Fragments Are Pseudomonas aeruginosa-Directed Antimicrobials Targeting Bacterial Replication. PLoS Pathog. 2015, 11, e1005159. [Google Scholar] [CrossRef]
- Wu, Z.; Hansmann, B.; Meyer-Hoffert, U.; Gläser, R.; Schröder, J.-M. Molecular identification and expression analysis of Filaggrin-2, a member of the S100 fused-type protein family. PLoS ONE 2009, 4, e5227. [Google Scholar] [CrossRef]
- Hansmann, B.; Ahrens, K.; Wu, Z.; Proksch, E.; Meyer-Hoffert, U.; Schröder, J. Murine filaggrin-2 is involved in epithelial barrier function and down-regulated in metabolically induced skin barrier dysfunction. Exp. Dermatol. 2012, 21, 271–276. [Google Scholar] [CrossRef]
- Bay, L.; Barnes, C.J.; Fritz, B.G.; Thorsen, J.; Restrup, M.E.M.; Rasmussen, L.; Sørensen, J.K.; Hesselvig, A.B.; Odgaard, A.; Hansen, A.J.; et al. Universal dermal microbiome in human skin. mBio 2020, 11, 10–1128. [Google Scholar] [CrossRef]
- Ryan, M.P.; Adley, C.C. Ralstonia spp.: Emerging global opportunistic pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Seo, K.S. Staphylococcus aureus Infection. Food Microbiol. Fundam. Front. 2023, 555–584. Available online: https://www.ncbi.nlm.nih.gov/books/NBK441868/ (accessed on 1 March 2024).
- Andrey, D.O.; Jousselin, A.; Villanueva, M.; Renzoni, A.; Monod, A.; Barras, C.; Rodriguez, N.; Kelley, W.L. Impact of the Regulators SigB, Rot, SarA and sarS on the Toxic Shock Tst Promoter and TSST-1 Expression in Staphylococcus aureus. PLoS ONE 2015, 10, e0135579. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Baldassarri, L.; Montanaro, L. In catheter infections by Staphylococcus epidermidis the intercellular adhesion (ica) locus is a molecular marker of the virulent slime-producing strains. J. Biomed. Mater. Res. 2001, 59, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Campoccia, D.; Ravaioli, S.; Montanaro, L. Polysaccharide intercellular adhesin in biofilm: Structural and regulatory aspects. Front. Cell. Infect. Microbiol. 2015, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Ballal, A.; Manna, A.C. Regulation of Superoxide Dismutase (sod) Genes by SarA in Staphylococcus aureus. J. Bacteriol. 2009, 191, 3301–3310. [Google Scholar] [CrossRef] [PubMed]
- Shtrichman, R.; E Samuel, C. The role of gamma interferon in antimicrobial immunity. Curr. Opin. Microbiol. 2001, 4, 251–259. [Google Scholar] [CrossRef]
- Zganiacz, A.; Santosuosso, M.; Wang, J.; Yang, T.; Chen, L.; Anzulovic, M.; Alexander, S.; Gicquel, B.; Wan, Y.; Bramson, J.; et al. TNF-α is a critical negative regulator of type 1 immune activation during intracellular bacterial infection. J. Clin. Investig. 2004, 113, 401–413. [Google Scholar] [CrossRef]
- Jayaraman, P.; Sada-Ovalle, I.; Nishimura, T.; Anderson, A.C.; Kuchroo, V.K.; Remold, H.G.; Behar, S.M. IL-1β Promotes Antimicrobial Immunity in Macrophages by Regulating TNFR Signaling and caspase-3 activation. J. Immunol. 2013, 190, 4196–4204. [Google Scholar] [CrossRef]
- Cooper, A.M. IL-17 and anti-bacterial immunity: Protection versus tissue damage. Eur. J. Immunol. 2009, 39, 649. [Google Scholar] [CrossRef]
- Spiegel, C.; Nogler, M.; Coraça-Huber, D.C. Sterilization Procedures for Titanium Alloy Surfaces Leads to Higher Expression of Biofilm-Related Staphylococcus aureus Genes. Antibiotics 2022, 11, 1647. [Google Scholar] [CrossRef]
- Anderson, J.M. Biocompatibility and Bioresponse to Biomaterials. In Principles of Regenerative Medicine; Academic Press: Cambridge, MA, USA, 2008; pp. 704–723. [Google Scholar]
- Anderson, J.M.; Rodriguez, A.; Chang, D.T. Foreign body reaction to biomaterials. In Seminars in Immunology; Academic Press: Cambridge, MA, USA, 2008; Volume 20, pp. 86–100. [Google Scholar]
- Rutala, W.A.; Weber, D.J. New disinfection and sterilization methods. Emerg. Infect. Dis. 2001, 7, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Zeitvogel, J.; Jokmin, N.; Rieker, S.; Klug, I.; Brandenberger, C.; Werfel, T. GATA3 regulates FLG and FLG2 expression in human primary keratinocytes. Sci. Rep. 2017, 7, 11847. [Google Scholar] [CrossRef]
- Mlitz, V.; Hussain, T.; Tschachler, E.; Eckhart, L. Filaggrin has evolved from an “S100 fused-type protein” (SFTP) gene present in a common ancestor of amphibians and mammals. Exp. Dermatol. 2017, 26, 955–957. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Ryu, J.Y.; Lee, J.S.; Choi, K.Y.; Chung, H.Y.; Cho, B.C.; Kim, K.; Lee, Y.J.; Jin, H.K.; Bae, J.-S.; et al. Effect of Breast Silicone Implant Topography on Bacterial Attachment and Growth: An In Vitro Study. In Vivo 2022, 36, 1703–1709. [Google Scholar] [CrossRef]
- Nam, S.Y.; Zhang, X.; Faruq, O.; Chien, P.N.; Dönmez, N.; Heo, C.Y. An Impact of Different Silicone Breast Implants on the Bacterial Attachment and Growth. J. Biomater. Nanobiotechnol. 2021, 12, 21–33. [Google Scholar] [CrossRef]
- Cohen, J.B.; Carroll, C.; Tenenbaum, M.M.; Myckatyn, T.M. Breast implant-associated infections: The role of the national surgical quality improvement program and the local microbiome. Plast. Reconstr. Surg. 2015, 136, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Palubicka, A.; Jaworski, R.; Wekwejt, M.; Swieczko-Zurek, B.; Pikula, M.; Jaskiewicz, J.; Zielinski, J. surgical site infection after breast surgery: A retrospective analysis of 5-year postoperative data from a single center in Poland. Medicina 2019, 55, 512. [Google Scholar] [CrossRef]
- Prantl, L.; Momeni, A.; Brebant, V.; Kuehlmann, B.; Heine, N.; Biermann, N.; Brix, E. Recommendations for the Use of Antibiotics in Primary and Secondary Esthetic Breast Surgery. Plast. Reconstr. Surg.-Glob. Open 2020, 8, e2590. [Google Scholar] [CrossRef]
- Federica, G.; Tommaso, F.; Alessia, C.; Agostino, C.; Florian, B.; Antonio, G.; Nicola, M.D.; Abdallah, R.; Carmela, S.; Lorenzo, S.; et al. Use of Antimicrobial Irrigation and Incidence of Capsular Contracture in Breast Augmentation and Immediate Implant-Based Breast Reconstruction. Aesthetic Plast. Surg. 2023, 47, 2345–2350. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Yang, S.; Zhang, Y. Peri-operative antibiotic prophylaxis does not reduce surgical site infection in breast cancer. Surg. Infect. 2020, 21, 268–274. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; Da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 1 March 2024).
- Barr, S.; Hill, E.; Bayat, A. Functional biocompatibility testing of silicone breast implants and a novel classification system based on surface roughness. J. Mech. Behav. Biomed. Mater. 2017, 75, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Atlan, M.; Kinney, B.M.; Perry, T.A. Intra- and Inter-Shell Roughness Variability of Breast Implant Surfaces. Aesthetic Surg. J. 2020, 40, NP324–NP326. [Google Scholar] [CrossRef] [PubMed]
- Atlan, M.; Bigerelle, M.; Larreta-Garde, V.; Hindié, M.; Hedén, P. Characterization of Breast Implant Surfaces, Shapes, and Biomechanics: A Comparison of High Cohesive Anatomically Shaped Textured Silicone, Breast Implants from Three Different Manufacturers. Aesthetic Plast. Surg. 2016, 40, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Grimus, V.; Coraça-Huber, D.C.; Steixner, S.J.M.; Nagl, M. Activity of N-Chlorotaurine against Long-Term Biofilms of Bacteria and Yeasts. Antibiotics 2021, 10, 891. [Google Scholar] [CrossRef] [PubMed]








| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Female sex | Severe coagulation disorder, representing a potential contraindication for the elective surgery |
| Age > 18 years | Rheumatic disease accompanied by obligatory intake of immunomodulating therapeutic agents |
| High-risk family history for breast and/or ovarian cancer and/or BRCA1/2 gene mutation carrier | Severe renal functional disorder: renal insufficiency status iv or v (estimated glomerulary filtration rate (gfr) < 30 mL/min) |
| Planned bilateral mastectomy with simultaneous breast reconstruction | Active hematological or oncological disease |
| HIV infection | |
| Hepatitis infection | |
| Pregnancy or breast-feeding | |
| Intake of anti-inflammatory drugs | |
| Carrier of silicone implants (e.g., gastric banding, mammary implants) | |
| Severe coagulation disorder, representing a potential contraindication for the elective surgery | |
| Rheumatic disease accompanied by obligatory intake of immunomodulating therapeutic agents |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schoberleitner, I.; Baier, L.; Lackner, M.; Zenz, L.-M.; Coraça-Huber, D.C.; Ullmer, W.; Damerum, A.; Faserl, K.; Sigl, S.; Steinkellner, T.; et al. Surface Topography, Microbial Adhesion, and Immune Responses in Silicone Mammary Implant-Associated Capsular Fibrosis. Int. J. Mol. Sci. 2024, 25, 3163. https://doi.org/10.3390/ijms25063163
Schoberleitner I, Baier L, Lackner M, Zenz L-M, Coraça-Huber DC, Ullmer W, Damerum A, Faserl K, Sigl S, Steinkellner T, et al. Surface Topography, Microbial Adhesion, and Immune Responses in Silicone Mammary Implant-Associated Capsular Fibrosis. International Journal of Molecular Sciences. 2024; 25(6):3163. https://doi.org/10.3390/ijms25063163
Chicago/Turabian StyleSchoberleitner, Ines, Leoni Baier, Michaela Lackner, Lisa-Maria Zenz, Débora C. Coraça-Huber, Wendy Ullmer, Annabelle Damerum, Klaus Faserl, Stephan Sigl, Theresia Steinkellner, and et al. 2024. "Surface Topography, Microbial Adhesion, and Immune Responses in Silicone Mammary Implant-Associated Capsular Fibrosis" International Journal of Molecular Sciences 25, no. 6: 3163. https://doi.org/10.3390/ijms25063163