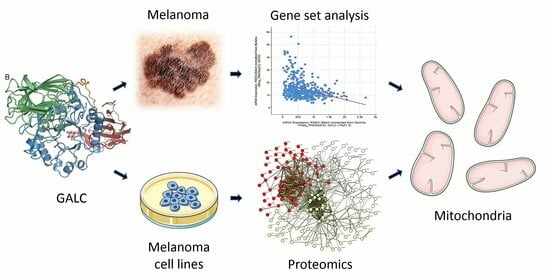
Proteomic Analysis Highlights the Impact of the Sphingolipid Metabolizing Enzyme β-Galactosylceramidase on Mitochondrial Plasticity in Human Melanoma
Abstract
:1. Introduction
2. Results
2.1. Negative Correlation between GALC and Nuclear-Encoded Mitochondrial Gene Expression in Human Melanoma
2.2. Proteomic Analysis of Downregulated Proteins in GALC-Overexpressing Melanoma Cells
3. Discussion
4. Materials and Methods
4.1. cBioPortal Data Mining
4.2. Proteomic Analysis
4.3. Categorization of Proteomic Data
4.4. Gene Set Enrichment Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Miller, A.J.; Mihm, M.C., Jr. Melanoma. N. Engl. J. Med. 2006, 355, 51–65. [Google Scholar] [CrossRef]
- Garbe, C.; Peris, K.; Hauschild, A.; Saiag, P.; Middleton, M.; Spatz, A.; Grob, J.J.; Malvehy, J.; Newton-Bishop, J.; Stratigos, A.; et al. Diagnosis and treatment of melanoma. European consensus-based interdisciplinary guideline—Update 2012. Eur. J. Cancer 2012, 48, 2375–2390. [Google Scholar] [CrossRef]
- Huang, C.; Radi, R.H.; Arbiser, J.L. Mitochondrial Metabolism in Melanoma. Cells 2021, 10, 3197. [Google Scholar] [CrossRef] [PubMed]
- Ottaviano, M.; Giunta, E.F.; Tortora, M.; Curvietto, M.; Attademo, L.; Bosso, D.; Cardalesi, C.; Rosanova, M.; De Placido, P.; Pietroluongo, E.; et al. BRAF Gene and Melanoma: Back to the Future. Int. J. Mol. Sci. 2021, 22, 3474. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef]
- Avagliano, A.; Fiume, G.; Pelagalli, A.; Sanita, G.; Ruocco, M.R.; Montagnani, S.; Arcucci, A. Metabolic Plasticity of Melanoma Cells and Their Crosstalk with Tumor Microenvironment. Front. Oncol. 2020, 10, 722. [Google Scholar] [CrossRef]
- Kumar, P.R.; Moore, J.A.; Bowles, K.M.; Rushworth, S.A.; Moncrieff, M.D. Mitochondrial oxidative phosphorylation in cutaneous melanoma. Br. J. Cancer 2021, 124, 115–123. [Google Scholar] [CrossRef]
- Du, F.; Yang, L.H.; Liu, J.; Wang, J.; Fan, L.; Duangmano, S.; Liu, H.; Liu, M.; Wang, J.; Zhong, X.; et al. The role of mitochondria in the resistance of melanoma to PD-1 inhibitors. J. Transl. Med. 2023, 21, 345. [Google Scholar] [CrossRef]
- Jamil, M.; Cowart, L.A. Sphingolipids in mitochondria-from function to disease. Front. Cell Dev. Biol. 2023, 11, 1302472. [Google Scholar] [CrossRef]
- Rodrigues, T.; Ferraz, L.S. Therapeutic potential of targeting mitochondrial dynamics in cancer. Biochem. Pharmacol. 2020, 182, 114282. [Google Scholar] [CrossRef]
- Belleri, M.; Chiodelli, P.; Corli, M.; Capra, M.; Presta, M. Oncosuppressive and oncogenic activity of the sphingolipid-metabolizing enzyme β-galactosylceramidase. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188675. [Google Scholar] [CrossRef]
- Won, J.S.; Singh, A.K.; Singh, I. Biochemical, cell biological, pathological, and therapeutic aspects of Krabbe’s disease. J. Neurosci. Res. 2016, 94, 990–1006. [Google Scholar] [CrossRef] [PubMed]
- Belleri, M.; Paganini, G.; Coltrini, D.; Ronca, R.; Zizioli, D.; Corsini, M.; Barbieri, A.; Grillo, E.; Calza, S.; Bresciani, R.; et al. β-Galactosylceramidase Promotes Melanoma Growth via Modulation of Ceramide Metabolism. Cancer Res. 2020, 80, 5011–5023. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Capoferri, D.; Chiodelli, P.; Corli, M.; Belleri, M.; Scalvini, E.; Mignani, L.; Guerra, J.; Grillo, E.; De Giorgis, V.; Manfredi, M.; et al. The Pro-Oncogenic Sphingolipid-Metabolizing Enzyme β-Galactosylceramidase Modulates the Proteomic Landscape in BRAF(V600E)-Mutated Human Melanoma Cells. Int. J. Mol. Sci. 2023, 24, 10555. [Google Scholar] [CrossRef]
- Capoferri, D.; Chiodelli, P.; Calza, S.; Manfredi, M.; Presta, M. Dataset: Impact of β-Galactosylceramidase Overexpression on the Protein Profile of Braf(V600E) Mutated Melanoma Cells. Data 2023, 8, 177. [Google Scholar] [CrossRef]
- Li, X.X.; Wang, Z.J.; Zheng, Y.; Guan, Y.F.; Yang, P.B.; Chen, X.; Peng, C.; He, J.P.; Ai, Y.L.; Wu, S.F.; et al. Nuclear Receptor Nur77 Facilitates Melanoma Cell Survival under Metabolic Stress by Protecting Fatty Acid Oxidation. Mol. Cell 2018, 69, 480–492.e7. [Google Scholar] [CrossRef] [PubMed]
- Aguera-Lorente, A.; Alonso-Pardavila, A.; Larrinaga, M.; Boyano, M.D.; Gonzalez, E.; Falcon-Perez, J.M.; Asumendi, A.; Apraiz, A. Small extracellular vesicle-based human melanocyte and melanoma signature. Pigment. Cell Melanoma Res. 2023, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ness, C.; Garred, Ø.; Eide, N.A.; Kumar, T.; Olstad, O.K.; Bærland, T.P.; Petrovski, G.; Moe, M.C.; Noer, A. Multicellular tumor spheroids of human uveal melanoma induce genes associated with anoikis resistance, lipogenesis, and SSXs. Mol. Vis. 2017, 23, 680–694. [Google Scholar]
- Liu, K.T.; Yeh, I.J.; Chou, S.K.; Yen, M.C.; Kuo, P.L. Regulatory mechanism of fatty acid-CoA metabolic enzymes under endoplasmic reticulum stress in lung cancer. Oncol. Rep. 2018, 40, 2674–2682. [Google Scholar] [CrossRef]
- Wu, X.; Zhao, J. Novel oxidative stress-related prognostic biomarkers for melanoma associated with tumor metastasis. Medicine 2021, 100, e24866. [Google Scholar] [CrossRef]
- Fujisawa, K.; Wakazaki, M.; Matsuzaki, A.; Matsumoto, T.; Yamamoto, N.; Noma, T.; Takami, T. Adenylate Kinase Isozyme 3 Regulates Mitochondrial Energy Metabolism and Knockout Alters HeLa Cell Metabolism. Int. J. Mol. Sci. 2022, 23, 4316. [Google Scholar] [CrossRef]
- Feng, S.; Zhu, G.; McConnell, M.; Deng, L.; Zhao, Q.; Wu, M.; Zhou, Q.; Wang, J.; Qi, J.; Li, Y.P.; et al. Silencing of atp6v1c1 prevents breast cancer growth and bone metastasis. Int. J. Biol. Sci. 2013, 9, 853–862. [Google Scholar] [CrossRef]
- Ayachi, O.; Barlin, M.; Broxtermann, P.N.; Kashkar, H.; Mauch, C.; Zigrino, P. The X-linked inhibitor of apoptosis protein (XIAP) is involved in melanoma invasion by regulating cell migration and survival. Cell. Oncol. 2019, 42, 319–329. [Google Scholar] [CrossRef]
- Han, D.; Zhu, W.; Chen, Y.; Wang, H. Parthenolide induces ROS-dependent cell death in human gastric cancer cell. Adv. Clin. Exp. Med. 2024, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pan, L.; Zhang, S.; Yang, Y.; Liang, J.; Ma, S.; Wu, Q. CISD2 promotes lung squamous carcinoma cell migration and invasion via the TGF-beta1-induced Smad2/3 signaling pathway. Clin. Transl. Oncol. 2023, 25, 3527–3540. [Google Scholar] [CrossRef]
- Llorca-Cardenosa, M.J.; Pena-Chilet, M.; Mayor, M.; Gomez-Fernandez, C.; Casado, B.; Martin-Gonzalez, M.; Carretero, G.; Lluch, A.; Martinez-Cadenas, C.; Ibarrola-Villava, M.; et al. Long telomere length and a TERT-CLPTM1 locus polymorphism association with melanoma risk. Eur. J. Cancer 2014, 50, 3168–3177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Li, Q.; Zhang, Y.; Li, N.; Rao, M.; Li, S.; Ai, Z.; Yan, S.; Tian, Z. COPS3 inhibition promotes cell proliferation blockage and anoikis via regulating PFKFB3 in osteosarcoma cancer cells. Eur. J. Pharmacol. 2023, 951, 175799. [Google Scholar] [CrossRef] [PubMed]
- Howell, M.C.; Green, R.; Khalil, R.; Foran, E.; Quarni, W.; Nair, R.; Stevens, S.; Grinchuk, A.; Hanna, A.; Mohapatra, S.; et al. Lung cancer cells survive epidermal growth factor receptor tyrosine kinase inhibitor exposure through upregulation of cholesterol synthesis. FASEB Bioadv. 2020, 2, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Hirata, A.S.; La Clair, J.J.; Jimenez, P.C.; Costa-Lotufo, L.V.; Fenical, W. Preclinical Development of Seriniquinones as Selective Dermcidin Modulators for the Treatment of Melanoma. Mar. Drugs 2022, 20, 301. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, W.; Zhang, J.; Liu, W.; Yin, J.; Shi, D.; Ma, W. A Novel Mitochondrial-Related Nuclear Gene Signature Predicts Overall Survival of Lung Adenocarcinoma Patients. Front. Cell Dev. Biol. 2021, 9, 740487. [Google Scholar] [CrossRef] [PubMed]
- Goswami, M.T.; VanDenBerg, K.R.; Han, S.; Wang, L.L.; Singh, B.; Weiss, T.; Barlow, M.; Kamberov, S.; Wilder-Romans, K.; Rhodes, D.R.; et al. Identification of TP53RK-Binding Protein (TPRKB) Dependency in TP53-Deficient Cancers. Mol. Cancer Res. 2019, 17, 1652–1664. [Google Scholar] [CrossRef] [PubMed]
- Schramm, K.; Iskar, M.; Statz, B.; Jager, N.; Haag, D.; Slabicki, M.; Pfister, S.M.; Zapatka, M.; Gronych, J.; Jones, D.T.W.; et al. DECIPHER pooled shRNA library screen identifies PP2A and FGFR signaling as potential therapeutic targets for diffuse intrinsic pontine gliomas. Neuro Oncol. 2019, 21, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Welinder, C.; Pawlowski, K.; Szasz, A.M.; Yakovleva, M.; Sugihara, Y.; Malm, J.; Jonsson, G.; Ingvar, C.; Lundgren, L.; Baldetorp, B.; et al. Correlation of histopathologic characteristics to protein expression and function in malignant melanoma. PLoS ONE 2017, 12, e0176167. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Yim, S.; Park, H. The cancer driver genes IDH1/2, JARID1C/KDM5C, and UTX/KDM6A: Crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism. Exp. Mol. Med. 2019, 51, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dubey, R.; Saini, N. STAT6 silencing up-regulates cholesterol synthesis via miR-197/FOXJ2 axis and induces ER stress-mediated apoptosis in lung cancer cells. Biochim. Biophys. Acta 2015, 1849, 32–43. [Google Scholar] [CrossRef]
- Zhang, K.; Liang, Y.; Zhang, W.; Zeng, N.; Tang, S.; Tian, R. KRT81 Knockdown Inhibits Malignant Progression of Melanoma Through Regulating Interleukin-8. DNA Cell Biol. 2021, 40, 1290–1297. [Google Scholar] [CrossRef]
- Liu, Q.; Lian, Q.; Song, Y.; Yang, S.; Jia, C.; Fang, J. Identification of LSM family members as potential chemoresistance predictive and therapeutic biomarkers for gastric cancer. Front. Oncol. 2023, 13, 1119945. [Google Scholar] [CrossRef]
- Cartron, P.F.; Petit, E.; Bellot, G.; Oliver, L.; Vallette, F.M. Metaxins 1 and 2, two proteins of the mitochondrial protein sorting and assembly machinery, are essential for Bak activation during TNF alpha triggered apoptosis. Cell Signal. 2014, 26, 1928–1934. [Google Scholar] [CrossRef]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Liu, J.; Kang, R.; Tang, D. Mitophagy Receptors in Tumor Biology. Front. Cell Dev. Biol. 2020, 8, 594203. [Google Scholar] [CrossRef]
- Zheng, B.; Chai, R.; Yu, X. Downregulation of NIT2 inhibits colon cancer cell proliferation and induces cell cycle arrest through the caspase-3 and PARP pathways. Int. J. Mol. Med. 2015, 35, 1317–1322. [Google Scholar] [CrossRef]
- Gameiro, P.A.; Laviolette, L.A.; Kelleher, J.K.; Iliopoulos, O.; Stephanopoulos, G. Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the tricarboxylic acid (TCA) cycle. J. Biol. Chem. 2013, 288, 12967–12977. [Google Scholar] [CrossRef] [PubMed]
- McDonald, S.L.; Edington, H.D.; Kirkwood, J.M.; Becker, D. Expression analysis of genes identified by molecular profiling of VGP melanomas and MGP melanoma-positive lymph nodes. Cancer Biol. Ther. 2004, 3, 110–120. [Google Scholar] [CrossRef]
- McGrail, K.; Granado-Martinez, P.; Esteve-Puig, R.; Garcia-Ortega, S.; Ding, Y.; Sanchez-Redondo, S.; Ferrer, B.; Hernandez-Losa, J.; Canals, F.; Manzano, A.; et al. BRAF activation by metabolic stress promotes glycolysis sensitizing NRAS(Q61)-mutated melanomas to targeted therapy. Nat. Commun. 2022, 13, 7113. [Google Scholar] [CrossRef]
- Zheng, J.F.; He, S.; Zeng, Z.; Gu, X.; Cai, L.; Qi, G. PMPCB Silencing Sensitizes HCC Tumor Cells to Sorafenib Therapy. Mol. Ther. 2019, 27, 1784–1795. [Google Scholar] [CrossRef]
- Bacchetti, T.; Salvolini, E.; Pompei, V.; Campagna, R.; Molinelli, E.; Brisigotti, V.; Togni, L.; Lucarini, G.; Sartini, D.; Campanati, A.; et al. Paraoxonase-2: A potential biomarker for skin cancer aggressiveness. Eur. J. Clin. Investig. 2021, 51, e13452. [Google Scholar] [CrossRef] [PubMed]
- Rebecca, V.W.; Nicastri, M.C.; Fennelly, C.; Chude, C.I.; Barber-Rotenberg, J.S.; Ronghe, A.; McAfee, Q.; McLaughlin, N.P.; Zhang, G.; Goldman, A.R.; et al. PPT1 Promotes Tumor Growth and Is the Molecular Target of Chloroquine Derivatives in Cancer. Cancer Discov. 2019, 9, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Li, L.; Wang, Z.; Pang, J.; Guan, X.; Yuan, Y.; Xia, Z.; Yi, W. A comprehensive analysis of the role of QPRT in breast cancer. Sci. Rep. 2023, 13, 15414. [Google Scholar] [CrossRef]
- Vivas-Garcia, Y.; Falletta, P.; Liebing, J.; Louphrasitthiphol, P.; Feng, Y.; Chauhan, J.; Scott, D.A.; Glodde, N.; Chocarro-Calvo, A.; Bonham, S.; et al. Lineage-Restricted Regulation of SCD and Fatty Acid Saturation by MITF Controls Melanoma Phenotypic Plasticity. Mol. Cell 2020, 77, 120–137.e9. [Google Scholar] [CrossRef]
- Bezawork-Geleta, A.; Rohlena, J.; Dong, L.; Pacak, K.; Neuzil, J. Mitochondrial Complex II: At the Crossroads. Trends Biochem. Sci. 2017, 42, 312–325. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Huang, L.; Huang, O.; Deng, Y.; Shi, X. A comprehensive analysis of SLC25A1 expression and its oncogenic role in pan-cancer. Discov. Oncol. 2023, 14, 207. [Google Scholar] [CrossRef] [PubMed]
- Kordass, T.; Weber, C.E.; Oswald, M.; Ast, V.; Bernhardt, M.; Novak, D.; Utikal, J.; Eichmuller, S.B.; Konig, R. SOX5 is involved in balanced MITF regulation in human melanoma cells. BMC Med. Genom. 2016, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Boese, A.C.; Kang, J.; Hwang, J.S.; Kim, J.; Eun, K.; Malin, C.M.; Magliocca, K.R.; Pan, C.; Jin, L.; Kang, S. Succinyl-CoA ligase ADP-forming subunit beta promotes stress granule assembly to regulate redox and drive cancer metastasis. Proc. Natl. Acad. Sci. USA 2023, 120, e2217332120. [Google Scholar] [CrossRef]
- McFarlane, R.J.; Wakeman, J.A. Translin-Trax: Considerations for Oncological Therapeutic Targeting. Trends Cancer 2020, 6, 450–453. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, Y.; Song, F.; Song, F.; Gao, C.; Liang, X.; Wang, F.; Chen, Z. URM1 Promoted Tumor Growth and Suppressed Apoptosis via the JNK Signaling Pathway in Hepatocellular Carcinoma. Onco Targets Ther. 2020, 13, 8011–8025. [Google Scholar] [CrossRef]
- Bizzozero, L.; Cazzato, D.; Cervia, D.; Assi, E.; Simbari, F.; Pagni, F.; De Palma, C.; Monno, A.; Verdelli, C.; Querini, P.R.; et al. Acid sphingomyelinase determines melanoma progression and metastatic behaviour via the microphtalmia-associated transcription factor signalling pathway. Cell Death Differ. 2014, 21, 507–520. [Google Scholar] [CrossRef] [PubMed]
- Montfort, A.; Bertrand, F.; Rochotte, J.; Gilhodes, J.; Filleron, T.; Milhes, J.; Dufau, C.; Imbert, C.; Riond, J.; Tosolini, M.; et al. Neutral Sphingomyelinase 2 Heightens Anti-Melanoma Immune Responses and Anti-PD-1 Therapy Efficacy. Cancer Immunol. Res. 2021, 9, 568–582. [Google Scholar] [CrossRef]
- Bilal, F.; Montfort, A.; Gilhodes, J.; Garcia, V.; Riond, J.; Carpentier, S.; Filleron, T.; Colacios, C.; Levade, T.; Daher, A.; et al. Sphingomyelin Synthase 1 (SMS1) Downregulation Is Associated With Sphingolipid Reprogramming and a Worse Prognosis in Melanoma. Front. Pharmacol. 2019, 10, 443. [Google Scholar] [CrossRef]
- Shirane, K.; Kuji, R.; Tareyanagi, C.; Sato, T.; Kobayashi, Y.; Furukawa, S.; Murata, T.; Kubota, S.; Ishikawa, Y.; Segawa, K.; et al. Gene expression levels of beta4-galactosyltransferase 5 correlate with the tumorigenic potentials of B16-F10 mouse melanoma cells. Glycobiology 2014, 24, 532–541. [Google Scholar] [CrossRef]
- Presta, M. β-Galactosylceramidase in cancer: Friend or foe? Trends Cancer 2021, 7, 974–977. [Google Scholar] [CrossRef] [PubMed]
- Rather, R.A.; Bhagat, M.; Singh, S.K. Oncogenic BRAF, endoplasmic reticulum stress, and autophagy: Crosstalk and therapeutic targets in cutaneous melanoma. Mutat. Res. Rev. Mutat. Res. 2020, 785, 108321. [Google Scholar] [CrossRef]
- Yoo, Y.A.; Kim, M.J.; Park, J.K.; Chung, Y.M.; Lee, J.H.; Chi, S.G.; Kim, J.S.; Yoo, Y.D. Mitochondrial ribosomal protein L41 suppresses cell growth in association with p53 and p27Kip1. Mol. Cell. Biol. 2005, 25, 6603–6616. [Google Scholar] [CrossRef]
- Nie, S.; Shi, Z.; Shi, M.; Li, H.; Qian, X.; Peng, C.; Ding, X.; Zhang, S.; Lv, Y.; Wang, L.; et al. PPARgamma/SOD2 Protects Against Mitochondrial ROS-Dependent Apoptosis via Inhibiting ATG4D-Mediated Mitophagy to Promote Pancreatic Cancer Proliferation. Front. Cell Dev. Biol. 2021, 9, 745554. [Google Scholar] [CrossRef]
- Li, Q.; Chu, Y.; Li, S.; Yu, L.; Deng, H.; Liao, C.; Liao, X.; Yang, C.; Qi, M.; Cheng, J.; et al. The oncoprotein MUC1 facilitates breast cancer progression by promoting Pink1-dependent mitophagy via ATAD3A destabilization. Cell Death Dis. 2022, 13, 899. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, H.T.; Suh, H.S. Combination therapy of BRAF inhibitors for advanced melanoma with BRAF V600 mutation: A systematic review and meta-analysis. J. Dermatol. Treat. 2018, 29, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology, C.; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]





| Protein | Gene | Biological Function |
|---|---|---|
| Acetyl-CoA Acyltransferase 2 | ACAA2 | 3-ketoacyl-CoA thiolase, mitochondrial. Catalyzes the last step of the mitochondrial beta-oxidation pathway. Its activity promotes melanoma cell survival and metastasis [17] |
| Acyl-CoA Dehydrogenase Medium Chain | ACADM | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial. Catalyzes the initial step of fatty acid β-oxidation. Its activity promotes melanoma cell survival and metastasis [17] |
| Aconitase 2 | ACO2 | Aconitate hydratase, mitochondrial. Catalyzes the isomerization of citrate to isocitrate via cis-aconitate in the TCA cycle. Enriched in melanoma small extracellular vesicles [18] |
| Acyl-CoA Thioesterase 1 | ACOT1 | Acyl-coenzyme A thioesterase 1, mitochondrial. Catalyzes the hydrolysis of acyl-CoAs, regulating intracellular levels of acyl-CoAs, free fatty acids, and coenzyme A. Associated with a lipogenic profile in uveal melanoma spheroids [19] |
| Acyl-CoA Thioesterase 13 | ACOT13 | Acyl-coenzyme A thioesterase 13, N-terminally processed, mitochondrial. High expression is associated with poor clinical outcomes in lung cancer [20] |
| Acyl-CoA Synthetase Long Chain Family Member 4 | ACSL4 | Long-chain-fatty-acid-CoA ligase 4, also mitochondrial. Catalyzes the conversion of long-chain fatty acids to acyl-CoA for both synthesis of cellular lipids and degradation via β-oxidation. Oxidative stress-related prognostic marker for melanoma metastasis [21] |
| Adenylate Kinase 3 | AK3 | GTP:AMP phosphotransferase AK3, mitochondrial. Involved in maintaining the homeostasis of cellular nucleotides by catalyzing the interconversion of nucleoside phosphates. AK3 knockout decreases proliferation and ATP levels in HeLa cells [22] |
| ATPase H+ Transporting V1 Subunit C1 | ATP6V1C1 | V-type proton ATPase subunit C 1; subunit of the peripheral V1 complex of vacuolar ATPase. V-ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells. ATP6V1C1 knockdown prevents breast cancer growth and bone metastasis [23] |
| Caspase 7 | CASP7 | Caspase-7 subunit p11. Involved in the activation cascade of caspases responsible for apoptosis execution. The caspase-7 inhibitor XIAP hampers melanoma invasion [24] |
| Catalase | CAT | Catalase. Protects cells from the toxic effects of hydrogen peroxide. Promotes melanoma growth. Inhibits ROS-induced cancer cell death [25] |
| CDGSH Iron Sulfur Domain 2 | CISD2 | CDGSH iron–sulfur domain-containing protein 2, also mitochondrial. Antagonizes BECN1-mediated cellular autophagy at the endoplasmic reticulum. CISD2 is aberrantly upregulated in malignant tumors [26] |
| Cleft Lip and Palate Transmembrane Protein 1-Like Protein | CLPTM1L | Cleft lip and palate transmembrane protein 1-like protein. TERT-CLPTM1 locus polymorphism is associated with melanoma risk [27] |
| COP9 Signalosome Subunit 3 | COPS3 | COP9 signalosome complex subunit 3. Component of the COP9 signalosome complex, an essential regulator of the ubiquitin conjugation pathway. The complex is also involved in phosphorylation of p53/TP53, c-jun/JUN, IκBα/NFKBIA, ITPK1, and IRF8/ICSBP. Acts as an oncogene in different cancers [28] |
| Cytochrome P450 Family 51 Subfamily A Member 1 | CYP51A1 | Lanosterol 14-alpha demethylase. A cytochrome P450 monooxygenase involved in sterol biosynthesis. Its inhibition decreases mitochondrial cholesterol and overcomes EGFR-TKI resistance in lung cancer cells [29] |
| Dermcidin | DCD | Survival-promoting peptide. DCD encodes the proteolysis-inducing factor core peptide (PIF-CP) and the skin antimicrobial peptide DCD-1. It may act as a pro-survival oncogene in various cancers, and it may represent a therapeutic target in melanoma [30] |
| G Elongation Factor Mitochondrial 1 | GFM1 | Elongation factor G, mitochondrial. Mitochondrial GTPase that catalyzes GTP-dependent ribosomal translocation during translation elongation. Associated with poor outcome in lung adenocarcinoma [31] |
| GON7 Subunit of KEOPS Complex | GON7 | EKC/KEOPS complex subunit GON7, mitochondrial. Component of the tRNA-modifying EKC/KEOPS complex that represents a potential therapeutic target in TP53-mutated cancer cells [32] |
| H3 Clustered Histone 1 | HIST1H3A | Histone H3.1. Core component of nucleosome. Recurrently mutated in diffuse intrinsic pontine gliomas [33] |
| Hexosaminidase Subunit Beta | HEXB | Hexosaminidase subunit beta chain A. Responsible for the degradation of GM2 gangliosides and other molecules containing terminal N-acetyl hexosamines. Hallmark of melanoma progression and poor survival [34] |
| Isocitrate Dehydrogenase (NADP(+)) 2 | IDH2 | Isocitrate dehydrogenase [NADP], mitochondrial. Plays a role in intermediary metabolism and energy production. Gain-of-function mutations drive tumor progression via D-2-hydroxyglutarate [35] |
| Isopentenyl-Diphosphate Delta Isomerase 1 | IDI1 | Isopentenyl-diphosphate Delta-isomerase 1. Involved in cholesterol synthesis. Upregulated following STAT6 silencing in lung cancer [36] |
| Keratin 86 | KRT86 | Keratin, type II cuticular Hb6. Knockdown of its KRT81 paralog inhibits melanoma progression [37] |
| LSM8 Homolog, U6 Small Nuclear RNA Associated | LSM8 | U6 snRNA-associated Sm-like protein LSm8. Plays a role in pre-mRNA splicing as a component of the U4/U6-U5 tri-snRNP complex. Upregulated in various human cancers and unfavorable biomarker in 5-FU-treated gastric cancer patients [38] |
| Metaxin 2 | MTX2 | Metaxin-2, mitochondrial. Involved in the transport of proteins into the mitochondrion as part of the VDAC2/Mtx1/Mtx2 multi-protein complex that incorporates the mitochondrial pro-apotptic protein Bak [39] |
| NEDD8 Activating Enzyme E1 Subunit 1 | NAE1 | NEDD8-activating enzyme E1 regulatory subunit. Regulatory subunit of the dimeric UBA3-NAE1 E1 enzyme. Necessary for cell cycle progression through the S m checkpoint. Inhibition of the neddylation pathway represses cancer cell growth [40] |
| Nipsnap Homolog 2 | NIPSNAP2 | Protein NipSnap homolog 2, mitochondrial. Modulator of mitochondrial calcium channels, it participates in mitophagy [41] |
| Nitrilase Family Member 2 | NIT2 | Omega-amidase NIT2, mitochondrial. A nitrilase that converts α-ketoglutaramate and α-ketosuccinamate to α-ketoglutarate and oxaloacetate, respectively. Its downregulation inhibits colon cancer cell proliferation [42] |
| Nicotinamide Nucleotide Transhydrogenase | NNT | NAD(P) transhydrogenase, mitochondrial. The transhydrogenation between NADH and NADP is coupled to respiration and ATP hydrolysis and functions as a proton pump across the membrane. Its knockdown activates glucose catabolism in melanoma cells [43] |
| NPC Intracellular Cholesterol Transporter 2 | NPC2 | NPC intracellular cholesterol transporter 2. Involved in the egress of cholesterol from the lysosomal compartment. Highly expressed in vertical growth phase melanomas and lymph node metastases [44] |
| Phosphofructokinase, Muscle | PFKM | ATP-dependent 6-phosphofructokinase, muscle type. Catalyzes the phosphorylation of D-fructose-6-phosphate to fructose-1,6-bisphosphate by ATP, the first committing step of glycolysis. Participates in the metabolic rewiring in NRAS-mutated melanoma [45] |
| Peptidase, Mitochondrial Processing Subunit Beta | PMPCB | Mitochondrial-processing peptidase subunit beta. Catalytic subunit of the essential mitochondrial processing protease that cleaves the mitochondrial sequence off newly imported precursor proteins. Contributes to tumor cell resistance against sorafenib [46] |
| Paraoxonase 2 | PON2 | Serum paraoxonase/arylesterase 2. Hydrolyzes lactones and several aromatic carboxylic acid esters. Exerts a protective role against ROS production within the mitochondrial respiratory chain. Its expression correlates with melanoma progression [47] |
| Palmitoyl-Protein Thioesterase 1 | PPT1 | Palmitoyl-protein thioesterase 1. Removes thioester-linked fatty acyl groups from modified cysteine residues in proteins or peptides during lysosomal degradation. Promotes tumor growth and is associated with poor prognosis in various cancers, including melanoma [48] |
| Quinolinate Phosphoribosyltransferase | QPRT | Nicotinate-nucleotide pyrophosphorylase [carboxylating]. Involved in the catabolism of quinolinic acid in the kynurenine pathway and mitochondrial dynamics. Modulates progression, metastasis, and invasion of breast cancer [49] |
| Stearoyl-CoA Desaturase | SCD | Acyl-CoA desaturase, mitochondrial. Stearyl-CoA desaturase catalyzes the insertion of a cis double bond into fatty acyl-CoA substrates. Regulates mitochondrial fatty acid oxidation and is required for MITF-mediated melanoma cell proliferation [50] |
| Succinate Dehydrogenase Complex Flavoprotein Subunit A | SDHA | Succinate dehydrogenase [ubiquinone], mitochondrial. Flavoprotein subunit of succinate dehydrogenase that functionally couples the TCA cycle with the electron transfer associated with OxPhos. Loss-of-function mutations increase the propensity for cellular transformation and tumor development [51] |
| Succinate Dehydrogenase Complex Iron Sulfur Subunit B | SDHB | Succinate dehydrogenase [ubiquinone], mitochondrial. Iron–sulfur protein subunit of succinate dehydrogenase. See SDHA [51] |
| Solute Carrier Family 25 Member 1 | SLC25A1 | Tricarboxylate transport protein, mitochondrial. Citrate transporter that mediates the exchange of mitochondrial citrate for cytosolic malate. Plays a pro-oncogenic role and may represent a prognostic biomarker in different cancers [52] |
| SRY-Box Transcription Factor 5 | SOX5 | Transcription factor SOX-5. Binds specifically to the DNA sequence 5′-AACAAT-3′. Highly expressed in melanoma cells, it inhibits MITF expression and is involved in melanocyte differentiation [53] |
| Succinate-CoA Ligase ADP-Forming Subunit Beta | SUCLA2 | Succinate--CoA ligase [ADP-forming] subunit beta, mitochondrial. ATP-specific succinyl-CoA synthetase functions in the TCA cycle, coupling the hydrolysis of succinyl-CoA to the synthesis of ATP. Its expression correlates with catalase levels and metastatic potential in lung and breast cancer patients [54] |
| Translin | TSN | Translin. DNA-binding protein that specifically recognizes consensus sequences at the breakpoint junctions in chromosomal translocations. Suppresses genome instability in Dicer-deficient cancers [55] |
| Ubiquitin Related Modifier 1 | URM1 | Ubiquitin-related modifier 1. Acts as a sulfur carrier required for 2-thiolation of various cytosolic tRNAs. Promotes tumor growth and suppresses apoptosis in hepatocellular carcinoma [56] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capoferri, D.; Mignani, L.; Manfredi, M.; Presta, M. Proteomic Analysis Highlights the Impact of the Sphingolipid Metabolizing Enzyme β-Galactosylceramidase on Mitochondrial Plasticity in Human Melanoma. Int. J. Mol. Sci. 2024, 25, 3062. https://doi.org/10.3390/ijms25053062
Capoferri D, Mignani L, Manfredi M, Presta M. Proteomic Analysis Highlights the Impact of the Sphingolipid Metabolizing Enzyme β-Galactosylceramidase on Mitochondrial Plasticity in Human Melanoma. International Journal of Molecular Sciences. 2024; 25(5):3062. https://doi.org/10.3390/ijms25053062
Chicago/Turabian StyleCapoferri, Davide, Luca Mignani, Marcello Manfredi, and Marco Presta. 2024. "Proteomic Analysis Highlights the Impact of the Sphingolipid Metabolizing Enzyme β-Galactosylceramidase on Mitochondrial Plasticity in Human Melanoma" International Journal of Molecular Sciences 25, no. 5: 3062. https://doi.org/10.3390/ijms25053062