1. Introduction
Glioblastoma multiforme (GBM) is a highly heterogeneous disease and is among the most common and aggressive forms of primary brain cancer. The molecular context of GBM has been categorized and successfully classified using multiple omics approaches. For instance, a study on The Cancer Genome Atlas (TCGA) published in 2010 discovered four subtypes of GBM: proneural, neural, classical, and mesenchymal. These subtypes are highly correlated with genetic modifications, such as tumor protein 53 (
TP53), epidermal growth factor receptor (
EGFR), and neurofibromatosis type 1 (
NF1) mutations [
1,
2]. GBM tumors with the mesenchymal signature have received considerable attention because they are more aggressive than those with other transcriptomic characteristics. Approximately a third of patients have the mesenchymal subtype of GBM, and these patients have a poor outcome. Compared with other subtypes of GBM tumors, mesenchymal tumors display higher levels of aggressiveness, angiogenic activity, hypoxia, inflammation, and multitherapy resistance [
3]. Thus, determining both standard-of-care and treatment approaches for patients with mesenchymal GBM is challenging. The identification of the key regulators of mesenchymal GBM and their downstream signaling pathways can enhance our understanding regarding this intricate transcriptional regulatory network and facilitate the development of effective treatment strategies.
GBM remains among the most fatal and difficult malignancies to treat, and less than 5% of patients survive for more than 5 years. Since 2005, patients with GBM undergoing maximally safe surgical resection have received temozolomide (TMZ), an alkylating drug approved by the Food and Drug Administration. In addition to causing DNA alkylation, TMZ causes oxidative stress, which is lethal for cancer cells [
4]. Although reactive oxygen species (ROS) are crucial second messengers in intracellular signaling pathways that moderately increase the oncogenic phenotype of cancer cells, they play a conflicting dual role. Excess ROS can damage proteins, lipids, and DNA, resulting in both apoptosis and ferroptosis [
5]. Furthermore, TMZ increases antioxidative activity, DNA repair mechanisms, and mitochondrial coupling, which promote tumor growth and chemoresistance [
6]. GBM recurrence is associated with shorter patient survival, resistance to TMZ, and increased Nuclear factor E2-related factor 2 (Nrf2)-targeted antioxidant system expression. Several mitochondria-related genes were differentially expressed between parental and resistant GBM cells, and among them, the superoxide dismutase 2 (
SOD2) gene substantially affected various factors, including cancer stemness, treatment resistance, and patient survival [
7].
SOD2 may regulate and neutralize a substantial portion of superoxide generated by the NADPH oxidase (NOX) family [
8]. The NOX family plays a protumorigenic role and promotes the mesenchymal features and carcinogenesis of several cancer cells [
9,
10]. Resistance to TMZ can be overcome by employing various approaches. Identifying the aberrant antioxidant system and treating it with pro-oxidants may be an effective strategy.
Ferroptosis is a form of regulated cell death governed by excess iron and characterized by ROS accumulation and lipid peroxidation, which cause membrane rupture. This nonapoptotic form of cell death was first described in 2012 by a study employing erastin to induce ferroptosis [
11]. Ferroptosis is effective in eliminating treatment-resistant tumor cells. Moreover, ferroptosis has been linked with drug resistance upon TMZ treatment. TMZ treatment has been known to accumulate mitochondrial and lipid ROS, increase labile iron pool, while depleting antioxidative capacity in glioma cells [
12,
13], thereby indicating ferroptosis induction as an alternative TMZ mechanism of action in glioblastoma. On the other hand, resistance to TMZ also closely connects with the way how glioma cells defend against the deleterious impact of a high ROS state and ferroptosis resilience [
7,
14]. Establishing this link may enable the use of ferroptosis inducers to treat gliomas. Nrf2 inhibition increased the sensitivity of glioma cells to TMZ [
15]. In addition, TMZ treatment promoted the generation of glutathione to confer resistance [
16]. Originally described as a multityrosine kinase inhibitor, sorafenib triggered ferroptosis and altered TMZ sensitivity which could be reversed through ROS elimination [
17]. These findings indicate that excessive ROS generation or pro-oxidant treatment may perturb TMZ sensitivity.
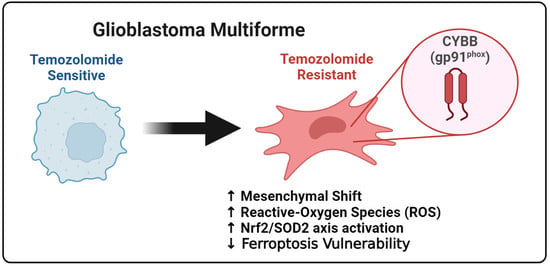
The identification of the possible link between chemoresistance and ferroptosis susceptibility, particularly in mesenchymal GBM, is helpful in determining the potential of ferroptosis to eliminate TMZ resistance. The findings of this study demonstrated that Cytochrome B-245 Beta Chain (CYBB), a major catalytic subunit of NOX, contributed to resistance upon TMZ treatment and altered the sensitivity of mesenchymal GBM to ferroptosis. Furthermore, aberrant CYBB activation in mesenchymal GBM led to the accumulation of mitochondrial ROS and activation of the compensatory Nrf2/SOD2 circuitry. This study determined the susceptibility of mesenchymal GBM to ferroptosis for overcoming TMZ resistance.
3. Discussion
Adult glioblastomas are the most prevalent and aggressive types of primary brain tumors. Despite the availability of intensive combinatorial chemotherapy, the survival rate for GBM remains low, with a median overall survival of approximately 1 year [
1]. Treatment-refractory and recurrent GBM is largely caused by heterogeneity within and between tumors. Because of their highly aggressive nature, GBM tumors harboring the mesenchymal signature have drawn considerable interest [
3]. EGFR and PDGFRA amplifications are prevalent in proneural and classical GBMs, respectively. The NF1 deficiency occurs in mesenchymal GBMs mainly through homozygous and hemizygous deletions. Loss of NF1 results in the infiltration of tumor-associated macrophages into the tumor microenvironment, causing mesenchymal transition and radioresistance to chemotherapy [
2,
3,
18]. In addition to suggesting a unique microenvironment orchestration, our study results suggest that mesenchymal GBMs contain certain deregulated oxidoreductases. We identified a NOX subunit, called CYBB, that can help in the detection but can also determine mesenchymal signature activation in GBM cells.
Glioblastoma with the mesenchymal subtype is highly invasive and expresses mesenchymal and proinflammatory genes. Cancer cells must retain mesenchymal and stemness properties to survive because stem-cell-like cells are resistant to treatment and promote tumor development. Our results revealed that CYBB expression was significantly associated with Nrf2 activation, CD44 overexpression, and N-cadherin overexpression in mesenchymal GBM cells. Nrf2 activation is commonly observed during mesenchymal GBM transformation [
19]. Thus, Nrf2 activation may serve as a surrogate marker for the prognosis of mesenchymal GBM. Furthermore, in this study, we noted that CYBB expression was associated with the aggressiveness of GBM. In gliomas exhibiting Nrf2 overactivity, CYBB might control the expression of antioxidant enzymes involved in drug detoxification and alleviate redox stress. Therefore, targeting events downstream of CYBB may provide an opportunity for clinicians to exploit this specific subpopulation of GBM to provide precision medicine.
The mesenchymal subtype of GBM is more aggressive and resistant to multiple therapies compared with other subtypes, implying the presence of metabolic abnormalities, such as altered redox homeostasis, due to an elevated metabolic rate and a harsh tumor microenvironment [
5]. NOX, a major source of intracellular superoxide, plays a significant role in regulating cellular redox signaling and homeostasis. NOX2, a member of the NOX family that was first identified to be expressed in myeloid cells, has been extensively studied in both normal and cancerous cells [
8]. The membrane-bound subunits CYBB (also called NOX2 or gp91phox, where phox refers to phagocyte oxidase) and p22phox (CYBA) are responsible for the catalytic core of oxidase. CYBB activation can result in the accumulation of ROS and contribute to the development of a resistant phenotype in acute myeloid leukemia (AML). AML with a high expression level of CYBB is associated with poor prognosis and chemoresistance [
20]. In this study, the aberrant expression of
CYBB and the antioxidant gene
SOD2 resulted in Nrf2 activation, and both these genes were differentially expressed in mesenchymal GBM. Therefore, understanding the interaction among CYBB, NRF2, and SOD2, which were markedly active in mesenchymal GBM, can explain the orchestration of pro-oxidative and antioxidative regulation that underlies treatment resistance in mesenchymal GBM.
Although ROS mediate key intracellular processes, such as carcinogenesis and tumor proliferation, their targeting causes collateral damage to host cells [
8]. ROS-mediated activation of Nrf2 is a crucial negative regulation step that can mitigate the detrimental effects of excessive ROS levels because NRF2 stimulates the expression of antioxidant enzymes. Neutrophil Cytosolic Factor 1 (NCF1) or p47-phox is physically bound to Nrf2, preventing the ubiquitination and activation of Nrf2 [
21]. However, our findings indicated that CYBB might physically interact with Nrf2 to promote its subsequent upregulation, thus causing mesenchymal shift, cancer stemness, and TMZ resistance in GBM. Cancer cells utilize Nrf2 transcriptional networks to counteract oxidative stress. Furthermore, tumor cells produce more antioxidative enzymes than normal cells, presumably to compensate for the toxicity of ROS generated by NOX [
8]. Because of the dual effects of ROS on both resistance and cell death, this study emphasizes that CYBB-mediated oxidative stress supports the mesenchymal features of GBM, activates Nrf2 to protect GBM cells from high ROS levels, and enhances the resistance of GBM cells to TMZ.
TMZ affects the bioenergetics and dynamics of mitochondria. Considerable changes in mitochondrial DNA and electron transport chain remodeling occur in response to TMZ-induced stress in GBM cells. The mitochondrial enzyme SOD2 was identified as a target of specificity protein 1 (Sp1); the increase in this protein in resistant cells is another mechanism underlying chemoresistance independent of methylguanine methyltransferase (MGMT) [
22]. The findings of this study indicated that Nrf2 activation upon aberrant CYBB expression regulates the transcription of SOD2 to promote TMZ resistance. This mechanism is vital to abrogate mitochondrial superoxide generation after exposure to TMZ. Other disease models have revealed that Nrf2 induces mitochondrial antioxidant enzymes, such as Sirt3 and SOD2, to maintain mitochondrial ROS homeostasis and protect neurons from oxidative damage [
23]. In addition, Nrf2 activation stimulates peroxisome proliferator-activated receptor coactivator 1α expression to protect cells against oxidative stress damage and enhance cisplatin resistance in ovarian cancer cells [
24]. Recent studies have identified SOD2 as a specific mitochondrial antioxidant system associated with TMZ resistance in mesenchymal GBM. Thus, the identification of alternative pro-oxidant approaches may help in overcoming acquired TMZ resistance.
Dixon et al. investigated how erastin selectively kills RAS-mutated tumor cells. Their research led to the discovery of ferroptosis, a cell death mechanism that considerably differs from apoptosis, necrosis, and autophagy in terms of cellular morphology, biochemistry features, and the key genes involved [
11]. Our results indicated that erastin-induced ferroptosis may kill TMZ-resistant GBM cells despite the mitochondrial antioxidant circuitry activity of Nrf2/SOD2. In response to TMZ treatment or ferroptosis inducers, such as erastin and TBHP, the suppression of
SOD2 expression resulted in the accumulation of mitochondrial superoxide. The superoxide radical is generated when an oxygen molecule receives an electron. By contrast, superoxide dismutase (SOD) is crucial for cellular defense against superoxide. Mitochondrial SOD2 converts superoxide into peroxide, which diffuses into the cytosol. Thus, SOD is essential to attenuate ferroptosis induction by inhibiting radical chain reactions.
The results of this study corroborate the previous reports that have highlighted the importance of SOD2 in buffering superoxide accumulation, as well as dictating sensitivity to chemotherapy and pro-ferroptosis agents [
7,
25,
26]. Specifically, our study showed that the depletion of SOD2 in TMZ-resistant GBM cells re-sensitized them to erastin-mediated ferroptosis, both in vitro and in vivo. Aside from that, inhibition of SOD2 increased mitochondrial ROS upon erastin or TMZ treatment. It has been known that the major form of ROS produced within mitochondria is superoxide [
8]. Previous studies have shown that decreased cellular antioxidant capacity, such as SOD2 levels, contributes to ferroptosis [
26,
27]. The antioxidative function of SOD2 is located within the mitochondrial matrix, where it converts superoxide anions produced by mitochondria during electron transport chain (ETC) to a less harmful radical, hydrogen peroxide (H
2O
2). As a result of the conversion of hydrogen peroxide to water by catalase, peroxidases, or peroxiredoxins, the cell is further preserved from deleterious oxidative stress. On the other hand, SOD2 deficiency leads to superoxide accumulation in the mitochondria, leading to oxidative stress. As both erastin and TMZ treatment produce ROS in cells [
13,
26], a depletion of SOD2 in the mitochondria can further increase superoxide levels. A higher superoxide level leads to increased lipid peroxidation in membranes and the occurrence of ferroptosis [
28]. In essence, mitochondrial oxidative stress resulting from SOD2 depletion can facilitate ferroptosis and increase the sensitivity of glioma cells to erastin or TMZ.
The susceptibility of TMZ-resistant GBM cells to ferroptosis may be explained by the interaction between CYBB and Nrf2/SOD2. In response to extrinsic treatment with drugs, such as TMZ, or intrinsically created ROS from NOX, Nrf2-mediated SOD2 transcription is a compensatory response. This study demonstrated that Nrf2/SOD2 prevented ferroptosis activation and excessive ROS production. Ferroptosis may be promoted by blocking Nrf2 or its downstream antioxidant genes through cotreatment with inducers, such as erastin and sorafenib [
29]. The suppression of
SOD2 can markedly increase ferroptosis sensitivity and suppress TMZ resistance in mesenchymal GBM. The findings of our animal study revealed that erastin-induced ferroptosis was highly effective in overcoming TMZ resistance. Targeting SOD2 may increase its efficacy in GBM. Thus, high CYBB expression may indicate not only mesenchymal features but also ferroptosis susceptibility in TMZ-resistant GBM. Erastin treatment may benefit mesenchymal GBM by exerting pro-oxidant effects and overcoming TMZ resistance. Because an erastin analog has not yet been tested in clinical trials, the use of tolerable drugs that inhibit SOD2 may promote ferroptosis while eliminating TMZ resistance in mesenchymal GBM; however, future studies on this topic are warranted.
4. Materials and Methods
4.1. Reanalysis of Single-Cell RNA Sequencing Dataset
We selected a representative single-cell transcriptome profiling dataset reported by Yu et al.; they used this dataset to explore molecular cross-talks between heterogeneous glioma cells and the microenvironment. We reanalyzed this dataset to determine the expression of the genes of interest in each cell cluster [
30]. The dataset by Yu et al. is deposited in the Genome Sequence Archive (GSA) database (accession number, HRA000179). In the GSA database, this dataset is named as the Chinese Glioma Genome Atlas (CGGA) and consists of approximately 8000 cells. After obtaining the file matrix, we used the Seurat package (version 4.0.6) in the R environment (version 4.0.1) to reconstruct the Seurat object. General preprocessing steps were performed, including the filtering of genes with no expression and reducing noise from the weakly expressed mitochondrial genome. Subsequently, the normalization and data scaling of the Seurat object were performed, followed by reducing the dimension and generation of cell clusters by employing the t-Distributed Stochastic Neighbor Embedding (t-SNE) module. The positive and negative markers of each cluster were generated and listed. The t-SNE plot, dot plot, and bar graph were constructed to present differences in the expression of the genes of interest between clusters.
4.2. Pathway Enrichment Analysis
To investigate key signaling pathways and biological processes perturbed in specific cell clusters, the Seurat module score was employed. The module score function was used to calculate the feature expression of each cell on the basis of selected gene set signatures or annotation. We used several functional annotations and curated gene sets following the previous gene signature to identify the subtype of GBM [
2]. Subsequently, we displayed the module score in an array of the t-SNE plot.
4.3. TCGA–GBM Dataset Acquisition
Bulk tumor RNA-sequencing data from TCGA for 172 patients with GBM, namely TCGA–GBM, were downloaded from the Xenabrowser portal, including their clinical information. The RNA-sequencing dataset is normalized and transformed by the web portal and presented in the “log2” format. The overall survival data of each patient were collected for survival analysis.
4.4. Cell Line Transcriptome Profiling
The Cancer Cell Line Encyclopedia dataset, which contains the RNA-sequencing profiles of almost all commercial glioma cell lines, was downloaded from the DepMap portal [
31]. Gene enrichment analysis of mesenchymal signatures and other functional annotations was performed using gene set variation analysis. A heatmap was generated to present the enrichment level of gene signatures in each cell line.
4.5. Identification of Differentially Expressed Genes
A normalized microarray dataset was prepared, and phenotypes for each sample were preset. The DESeq2 package (version 3.52.2) was used to count the fold-change difference and significance level for each gene. Genes with a |log2 fold-Change| of ≥1 and a p value of <0.05 were considered as differentially expressed genes (DEGs). DEGs from each dataset were presented in a Venn diagram to determine shared and common DEGs between the phenotypes of interest.
4.6. Immunohistochemical Tissue Staining
We detected the expression levels of CYBB protein and mesenchymal markers, namely vimentin (Vim), CD44, and N-cadherin, in 65 GBM tissue specimens from the TMU–SHH GBM cohort by performing immunohistochemical (IHC) analysis. Formalin-fixed, paraffin-embedded tissue sections were placed on coated glass slides. IHC analysis was performed in accordance with the standard protocol involving antigen retrieval, primary antibody incubation, secondary chromogen addition, and hematoxylin counterstaining. Rabbit anti-CYBB monoclonal antibody was used at a dilution of 1:200. Images were captured using a microscope (Leica Microsystems, Wetzlar, Germany). Immunostaining expression of each tissue specimen was scored using the IHC Q or quick score on the basis of the intensity and extent of expression. The Q-score method has been described previously. The intensity was scored as follows: 0 = negative, 1 = weak, 2 = medium, and 3 = high. The extent of expression was evaluated as the percentage of the positively stained area compared with the entire tumor area. The final IHC Q score was determined by quantifying the intensity score and positivity percentage. The minimum and maximum IHC scores were 0 and 300, respectively. The tissue specimens were categorized into high and low CYBB expression groups by using the median Q score of the entire cohort as the cutoff. The association of the IHC Q score with certain clinical parameters of each patient was evaluated.
4.7. Cell Line Culture
The human GBM cell line U87MG was purchased from the American Type Culture Collection (Manassas, VA, USA). U87MG cells primarily possess the mesenchymal phenotype [
32]. The cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Invitrogen). The cells were grown at 37 °C in an incubator with 5% humidified CO
2. Cell passage was performed when the cells had almost reached full confluence. The culture medium was replaced with fresh medium every 72 h before the administration of TMZ or any other treatment.
4.8. Generation of TMZ-Resistant Cells
TMZ-resistant U87MGR cells were generated by treating them at increasing doses (up to 150 µM) of TMZ for 2 weeks, followed by maintenance treatment with 100 µM TMZ in vitro in accordance with a previous study [
33]. Chemosensitivity was evaluated by performing the sulforhodamine B viability assay, and the corresponding drug–response curve was plotted to indicate the generation of TMZ resistance in U87MGR cells.
4.9. shRNA-Mediated Knockdown
The lentiviral CYBB shRNA (Cat#: TL313616V) and SOD2 shRNA (Cat#: TL309190V) were obtained from Origene (Rockville, MD, USA) and used to suppress the expression of the corresponding genes in accordance with the manufacturer’s protocol. The nontarget sequence (Scr shRNA, Origene Cat#: TR30021V) was used as control. Transfection was performed for 24 h by using Lipofectamine 2000 in accordance with the manufacturer’s instructions. The knockdown efficiency was verified using immunoblotting or quantitative polymerase chain reaction.
4.10. Immunofluorescence Staining
Immunofluorescence staining was performed to determine changes in particular markers under specific conditions in GBM cells. Prior to staining, either scramble or shCYBB U87MG-R cells were cultured in six-well plates and treated with 100 µM TMZ for 24 h or left untreated. Subsequently, the cells were plated in six-well chamber slides and permeabilized by treating them with 0.1% Triton X-100 in 0.01 M phosphate-buffered saline (PBS, pH 7.4). The cells were washed twice in PBS containing 1% bovine serum albumin and stained with rabbit anti-Nrf2 (dilution 1:100, cat. #7631, Cell Signaling, Danvers, MA, USA) overnight. The stained cells were washed three times, resuspended in a mounting medium, and fixated onto coverslips. Then, 4′,6-diamidino-2-phenylindole (DAPI) was used for nuclear staining. Photographs were obtained using a Leica spectral confocal fluorescence imaging system.
4.11. Cell Death Analysis
Each resistant or parental cell line, with either CYBB or SOD2 knockdown, was exposed to either TMZ, erastin, or tert-butyl hydroperoxide (TBHP) with or without cotreatment with ferroptosis inhibitors for approximately 24 h. Subsequently, the cells were harvested, washed with PBS, and fixed with ice-cold 70% ethanol at −20 °C for 30 min. Then, cells were incubated with 5 µg/mL RNase for 30 min at room temperature and stained with propidium iodide (PI, 5 µg/mL) for 1 h. Dead cells were stained with PI and examined through flow cytometry.
4.12. Tumorsphere Study
The cells were seeded in serum-free low-adhesion culture plates containing RPMI1640 with B27 supplement (Invitrogen), 20 ng/mL EGF, and 20 ng/mL basic FGF (stem cell medium; PeproTech, Rocky Hill, NJ, USA) for approximately 14 days to allow the formation of tumorspheres. Spheres were counted under a microscope. The tumorsphere formation efficiency was calculated as the ratio of the number of tumorspheres formed to the seeded adherent cell number.
4.13. Colony Formation Study
To observe the generation of tumor colonies, approximately 2500 GBM cells/cm2 were suspended in 0.3% agarose containing the tumorsphere medium (StemCell Technologies, Vancouver, BC, Canada) and layered on a 0.8% agar base layer. The culture was covered with 0.5 mL of tumorsphere medium and cultured for 14 days. For quantification, the wells were observed under a microscope, and tumor colonies were analyzed using ImageJ software.
4.14. Invasion and Migration Study
To evaluate the migration and invasion capabilities of cancer cells, Transwell migration and invasion assays were performed. Briefly, U87MG or U87MG-R cells were incubated in six-well plates for 24 h until they reached full confluency. Matrigel precoating was required for invasion studies before seeding with 1 × 105 cells, whereas migration studies did not require precoating the membrane. In two chambers, different concentrations of BSA were added, with the upper chamber containing RPMI with 2% and the lower chamber containing 20% FBS. Incubation was conducted for 48 h, either with or without treatment. Cells that penetrated membranes were carefully removed, and those that migrated or invaded were fixed in ethanol and stained with crystal violet. Under a microscope, five randomly selected fields were examined, and representative images were captured to determine the number of migrated or invaded cells.
4.15. Mitochondrial ROS Study
The superoxide indicator MitoSOX red (Invitrogen, Karlsruhe, Germany) was used to examine mitochondrial ROS production. MitoTracker Green FM (Invitrogen, Karlsruhe, Germany) was used to detect mitochondria. Prior to staining, cells were grown in six-well plates and treated with 100 µM TMZ, 150 µM TBHP, 10 µM Erastin, or 2 µM SRS11-92 for 12 h or left untreated. The cells were then detached, the medium was replaced, and approximately 30,000 cells/well were regrown on eight-well slides. MitoSOX (1 µM) and MitoTracker Green FM (150 nM) were applied for 20 min to U87MG and U87MG-R cells. Hoechst dye was used to counterstain the cells. The cells were fixed with 4% paraformaldehyde for 20 min at room temperature. A fluorescence microscope was used to obtain images.
4.16. Reporter Luciferase Assay
The pGL3-SOD2 promoter Luc reporter gene construct was generated using a plasmid pGL3-basic (Promega, Madison, WI, USA) containing the luciferase gene and a human SOD2 promoter fragment as described previously [
34,
35] to evaluate the transcriptional activity of the SOD2 promoter. The SOD2 promoter region spanning from −3340 to +260 nucleotides was cloned into a pGL3 basic vector. The putative Nrf2 or antioxidant response element binding site was located at +65 to +76 of the cloned SOD2 promoter. The reporter gene construct was transfected into U87MG-R cells by using Lipofectamine 2000 in accordance with the manufacturer’s protocol. Eighteen hours after the transfection, the cells were exposed to TBHP or TMZ or GSK2795039 (NOX inhibitor) for twenty-four hours. Luciferase activity was determined using a dual-luciferase reporter assay kit (Promega, Madison, WI, USA).
4.17. Tumor Xenograft Animal Study
Animal experiments were performed using 5-to-6-week-old female BALB/C nude mice obtained from BioLASCO (Taipei City, Taiwan). Animals were maintained in an animal center at 22–28 °C under a 12 h diurnal cycle and provided unrestricted access to food and water. Using Matrigel mixture (Becton Dickinson, Bedford, MA, USA), we subcutaneously injected 100 µL solution containing 1 × 106 scrambled or shSOD2 U87MG-R cells into the left flank of each mouse as part of in vivo studies (The IACUC Approval Number LAC-2021-0578). Imidazole ketone erastin (IKE) was used as an erastin analog to induce ferroptosis in mice harboring either scramble or shSOD2 U87MG-R xenografts. A total of four groups were formed by randomly dividing the mice: control group consisting of shScramble xenografts treated with vehicle (n = 5), shScramble xenografts treated with IKE (n = 5), shSOD2 xenografts treated with vehicle (n = 5), and shSOD2 xenografts treated with IKE (n = 5). Drug treatment was started after the tumor volume reached approximately 100 mm3 or 10 days after the injection of the tumor xenograft. Depending on the treatment group, vehicles (5 mg/kg/day) or IKE (25 mg/kg/day) were administered intraperitoneally. Treatment was administered intraperitoneally for 3 weeks, and tumor growth was observed for 6 weeks after treatment began. Tumor growth was evaluated every 2 weeks by using Vernier calipers, and tumor volume (V) was calculated using the following formula: V = 0.5 × [long diameter × short diameter2]. Tumor mass was collected in week 6 after mice were sacrificed and used in further experiments.
4.18. Statistical Analysis
Numerical variables are presented as the mean and standard deviation, whereas categorical variables are expressed as the frequency and percentage. An association between categorical variables was assessed using the chi-square test. Student’s t-test was used to compare continuous variables within two groups, and one-way analysis of variance (ANOVA) was used to evaluate discrepancies among more than two groups. Two-way ANOVA was used to determine the differential responses of numerical variables by two independent variables. Kaplan–Meier analysis was performed to estimate survival curves, and the log-rank test was performed to calculate p values. Cox regression was performed using univariate and multivariate methods to assess the effect of the risk group on the survival of patients with GBM. Statistical significance was set at p < 0.05. Data analysis was performed using GraphPad Prism 6.0 (San Diego, CA, USA) and SPSS version 21.0 (IBM, Armonk, NY, USA).