Inhibition of ROS-Scavenging Enzyme System Is a Key Event in Tomato Genetic Resistance against Root-Knot Nematodes
Abstract
:1. Introduction
2. Results
2.1. Bioassays of Tomato–RKN Interactions
2.2. ROS-Scavenging Enzyme Assays in Root Crude Extracts Collected from the Earliest Stages of Tomato–RKN Incompatible Interactions
2.3. CAT Inhibition as a Marker of Tomato Resistance to RKNs
2.4. Expression of Antioxidant and Cell Death Promoting Genes Involved in Incompatible and Compatible RKN–Tomato Interactions
2.5. Biochemical Mechanisms of CAT Inhibition in Tomato Resistance to RKNs
Purification and Characterization of CAT Isozymes from a Tomato Resistant Isoline
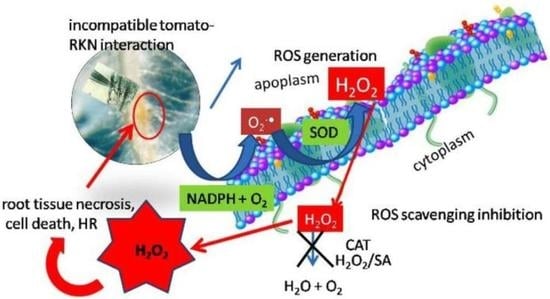
2.6. Generation of H2O2 in Tomato Resistance to Nematodes
3. Discussion
4. Materials and Methods
4.1. Procedures to Realize Compatible and Incompatible Tomato–RKN Interactions
4.2. Nematode Inoculations and Infection Level Determination
4.3. Extraction of Cellular Fractions from Root Samples
4.4. Purification of CAT from Soluble Fractions of Root Samples
4.5. Proteins, Phenols, and Enzyme Spectrophotometric Assays
- -
- An amount of 0.05 M phosphate buffer (pH 6.0), 5 mM guaiacol or 50 µM syringaldazine, 2 mM H2O2, and 10–20 µL microsome suspension.
- -
- An amount of 0.1 M Tris-HC1 buffer (pH 7.6), 0.35 mM PPD/4.5 mM PC, 2 mM H2O2, and 10–20 µL microsome suspension.
- -
- One unit of isoperoxidase activity indicated difference in absorbance in Arbitrary Units (∆AUx) min−1 mg−1 prot.
4.6. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time Polymerase Chain Reaction
- F: GTTTGCTTGCACACGGTTTA/R: CGTCGTTGGTGGATACCTCT for GPX,
- F:TGCTCCAAAGTGTGCTCATC/R:TTGCATCCTCCTCTGAAACC for CAT 2
- F: TGACCAACACAGGAACAGGA/R: GCCCAATCCATTAGTGTCCA for PR-4b
- F: CAGCAGATGTGGATCTCAAA/R: CTGTGGACAATGGAAGGAC for ACT-7
4.7. Experimental Design and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, J.T.; Haegeman, A.; Danchin, E.G.; Gaur, H.S.; Helder, J.; Jones, M.G.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.; et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Molinari, S. Can the plant immune system be activated against animal parasites such as nematodes? Nematology 2020, 22, 1–12. [Google Scholar] [CrossRef]
- Sato, K.; Kadota, Y.; Shirazu, K. Plant immune responses to parasitic nematodes. Front. Plant Sci. 2019, 10, 1165. [Google Scholar] [CrossRef]
- Milligan, S.B.; Bodeau, J.; Yaghoobi, J.; Kaloshian, I.; Zabel, P.; Williamson, V.M. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 1998, 10, 1307–1319. [Google Scholar] [CrossRef]
- Rossi, M.; Goggin, F.L.; Milligan, S.B.; Kaloshian, I.; Ullman, D.E.; Williamson, V.M. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 1998, 95, 9750–9754. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Molinari, S. Molecular aspects of plant-nematode interaction. Nematol. Medit. 1996, 24, 139–154. [Google Scholar]
- Kadota, Y.; Shirasu, K.; Zipfel, C. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar] [CrossRef]
- Dropkin, V.H. The necrotic reaction of tomatoes and other hosts resistant to Meloidogyne: Reversal by temperature. Phytopathology 1969, 59, 1632–1637. [Google Scholar]
- Branch, C.; Hwang, C.F.; Navarre, D.A.; Williamson, V.M. Salicylic acid is part of the Mi-1-mediated defense response to root-knot nematode in tomato. Mol. Plant-Microbe Interact. 2004, 17, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Molinari, S.; Loffredo, E. The role of salicylic acid in defense response of tomato to root-knot nematodes. Physiol. Mol. Plant Pathol. 2006, 68, 69–78. [Google Scholar] [CrossRef]
- Molinari, S. New developments in understanding the role of salicylic acid in plant defence. CAB Rev. 2007, 2, 1–10. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting mechanisms of defense aganst biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Vasyukova, N.I.; Pridvorova, S.M.; Gerasimova, N.G.; Chalenko, G.I.; Ozeretskovskaya, O.L.; Udalova, Z.V.; Zinov’eva, S.V. The involvement of phenylalanine ammonia-lyase and salicylic acid in the induction of resistance of tomato plants infested with gall nematode Meloidogyne incognita. Doklady Biol. Sci. 2007, 416, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Molinari, S.; Fanelli, E.; Leonetti, P. Expression of tomato salicylic acid (SA)-responsive pathogenesis-related genes in Mi-1-mediated and SA-induced resistance to root-knot nematodes. Mol. Plant Pathol. 2014, 15, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Tenhaken, R.; Rübel, C. Salicylic acid is needed in hypersensitive cell death in soybean but does not act as a catalase inhibitor. Plant Physiol. 1997, 115, 291–298. [Google Scholar] [CrossRef]
- Yeom, S.-I.; Baek, H.-K.; Oh, S.-K.; Kang, W.H.; Lee, S.J.; Lee, J.M.; Seo, E.; Rose, J.K.; Kim, B.D.; Choi, D. Use of a secretion trap screen in pepper following Phytophthora capsici infection reveals novel functions of secreted plant proteins in modulating cell death. Mol. Plant-Microbe Interact. 2011, 24, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Molinari, S.; Leonetti, P. Bio-control agents activate plant immune response and prime susceptible tomato against root-knot nematodes. PLoS ONE 2019, 14, e0213230. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Lv, D.; Wang, P.; Wang, X.C.; Chen, J.; Miao, C.; Song, C.P. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 2006, 18, 2749–2766. [Google Scholar] [CrossRef]
- Hwang, I.S.; Choi, D.S.; Kim, N.H.; Kim, D.S.; Hwang, B.K. Pathogenesis-related protein 4b interacts with leucine-rich repeat protein 1 to suppress PR4b-triggered cell death and defense response in pepper. Plant J. 2014, 77, 521–533. [Google Scholar] [CrossRef]
- Dincer, A.; Aydemir, T. Purification and characterization of catalase from chard (Beta vulgaris var. cicla). J. Enzyme Inhibit. 2001, 16, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Doke, N. NADPH-dependent O2− generation in membrane fraction isolated from wounded potato tubers inoculated with Phytophtorainfestans. Physiol. Plant Pathol. 1985, 27, 311–322. [Google Scholar] [CrossRef]
- Elthon, T.E.; Heupel, A. Formation of hydrogen peroxide by isolated cell walls from horseradish (Armoracia lapathifolia Gibil.). Planta 1976, 130, 175–180. [Google Scholar]
- Nombela, G.; Williamson, V.M.; Muniz, M. The root-knot nematode resistance gene Mi-1.2of tomato is responsible for resistance against the whitefly Bemisiatabaci. Mol. Plant-Microbe Interact. 2003, 16, 645–649. [Google Scholar] [CrossRef]
- Paulson, R.E.; Webster, J.M. Ultrastructure of the hypersensitive reaction in roots of tomato Lycopersicon esculentum L., to infection by the root-knot nematode, Meloidogyne incognita. Physiol. Plant Pathol. 1972, 2, 227–232. [Google Scholar] [CrossRef]
- Hwang, C.-F.; Bhakta, A.V.; Truesdell, G.M.; Pudio, W.M.; Williamson, V.M. Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 2000, 12, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nature Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef]
- Schaff, J.E.; Nielsen, D.M.; Smith, C.P.; Scholl, E.H.; Bird, D.M. Comprehensive transcriptome profiling in tomato reveals a role for glycosyltransferase in Mi-mediated nematode resistance. Plant Physiol. 2007, 144, 1079–1092. [Google Scholar] [CrossRef]
- Melillo, M.T.; Leonetti, P.; Bongiovanni, M.; Castagnone-Sereno, P.; Bleve-Zacheo, T. Modulation of reactive oxygen species activities and H2O2 accumulation during compatible and incompatible tomato-root knot nematode interactions. New Phytol. 2006, 170, 501–512. [Google Scholar] [CrossRef]
- Crane, F.L.; Sun, I.L.; Clark, M.G.; Grebing, C.; Low, H. Trans-membrane redox system in growth and development. Biochem. Biophys. Acta 1985, 811, 233–264. [Google Scholar] [PubMed]
- Schwacke, R.; Hager, A. Fungal elicitors induce a transient release of active oxygen species from cultured spruce cells that is dependent on Ca2+ and protein-kinase activity. Planta 1992, 187, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Molinari, S. Regulation of the superoxide anion generation in tomato roots susceptible and resistant to nematodes. Nematolmedit 1994, 22, 69–74. [Google Scholar]
- Niebel, A.; Heungens, K.; Barthels, N.; Inzé, D.; Van Montagu, M.; Gheysen, G. Characterization of a pathogen-induced potato catalase and its systemic expression upon nematode and bacterial infection. Mol. Plant-Microbe Interact. 1995, 8, 371–378. [Google Scholar] [CrossRef]
- Durner, J.; Klessig, D.F. Salicylic acid is a modulator of tobacco and mammalian catalases. J. Biol. Chem. 1996, 271, 28492–28501. [Google Scholar] [CrossRef]
- Li, Q.; Xie, Q.G.; Smith-Becker, J.; Navarre, D.A.; Kaloshian, I. Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling cascades. Mol. Plant Microbe Interact. 2006, 19, 655–664. [Google Scholar] [CrossRef]
- Ganesan, V.; Thomas, G. Salicylic acid response in rice: Influence of salicylic acid on H2O2 accumulation and oxidative stress. Plant Sci. 2001, 160, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Enyedi, A.J.; Yalpani, N.; Silverman, P.; Raskin, I. Signal molecules in systemic plant resistance to pathogens and pests. Cell 1992, 70, 879–886. [Google Scholar] [CrossRef]
- Mazel, A.; Levine, A. Induction of cell death in Arabidopsis by superoxide in combination with salicylic acid or protein synthesis inhibitors. Free Radic. Biol. Med. 2001, 30, 98–106. [Google Scholar] [CrossRef]
- Molinari, S. Immune responses induced by salicylic and jasmonic acids against plant parasites. In Salicylic Acid and Jasmonic Acid: Biosynthesis, Functions and Role in Plant Development, Plant Science Research and Practices; Santos, P., Ed.; Nova Science Publishers, Inc.: New York, NY, USA, 2015; pp. 1–35. [Google Scholar]
- Ryals, J.A.; Neuenschwander, U.H.; Willits, N.G.; Molina, A.; Steiner, H.Y.; Hunt, M.D. Systemic acquired resistance. Plant Cell 1996, 8, 1809–1819. [Google Scholar] [CrossRef]
- Norman, C.; Howell, K.A.; Millar, A.H.; Whelan, J.M.; Day, D.A. Salicylic acid is an uncoupler and inhibitor of mitochondrial electron transport. Plant Physiol. 2004, 134, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Molinari, S.; Zacheo, G.; Bleve-Zacheo, T. Effects of nematode infestation on mitochondria isolated from susceptible and resistant tomato roots. Physiol. Mol. Plant Pathol. 1990, 37, 27–37. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Chance, B.; Mahley, A.C. Assay of catalases and peroxidases. In Methods in Enzimology; Colowick, S.P., Kaplan, N.O., Eds.; Academic Press: New York, NY, USA, 1955; Volume 2, pp. 764–775. [Google Scholar]
- Furusawa, I.; Tanaka, K.; Thantuong, P.; Miziguchi, A.; Yazaki, M.; Asada, K. Paraquat resistant tobacco calluses with enhanced superoxide dismutase activity. Plant Cell Physiol. 1984, 25, 1247–1254. [Google Scholar]
- Gerbling, K.P.; Kelly, G.J.; Fisher, K.H.; Latzko, E. Partial purification and properties of soluble ascorbate peroxidase from pea leaves. J. Plant Pathol. 1984, 115, 59–67. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Baker, C.J.; Orlandi, E.W. Active oxygen in plant pathogenesis. Annu. Rev. Phytopathol. 1995, 33, 299–321. [Google Scholar] [CrossRef] [PubMed]
- Molinari, S. Effect of paraquat on tomato roots cultured in vitro susceptible and resistant to nematodes. Plant Cell Physiol. 1991, 32, 1129–1135. [Google Scholar]
- Sawhney, R.; Webster, J.M. The influence of some metabolic inhibitors on the response of susceptible/resistant cultivars of tomato to Meloidogyne incognita. Nematologica 1979, 25, 86–93. [Google Scholar] [CrossRef]
- Edenss, R.M.; Anand, S.C.; Bolla, R.I. Enzymes of the phenylpropanoid pathway in soybean infected with Meloidogyne incognita or Heterodera glycines. J. Nematol. 1995, 27, 292–303. [Google Scholar]








| Incompatible Interactions | Compatible Interactions | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| cv. Motelle | cv. VFN8 | cv. Moneymaker | cv. Roma VF | cv. VFN8 | ||||||
| cntr | + MifieldV | cntr | + MifieldV | cntr | + MifieldV | cntr | + MifieldV | cntr | + SM2V | |
| CFs | ||||||||||
| CAT | 27.4 ± 6.3 | 17.8 ± 4.8 * | 12.5 ± 1.1 | 6.0 ± 1.2 * | 9.1 ± 1.2 | 9.6 ± 1.1 | 8.9 ± 3.1 | 7.8 ± 1.5 | 19.2 ± 4.1 | 21.1 ± 1.6 |
| PHE | 70 ± 40 | 40 ± 20 * | 30 ± 10 | 10 ± 10 * | 40 ± 10 | 50 ± 10 | 40 ± 30 | 40 ± 20 | ||
| MPFs | ||||||||||
| CAT | 34.2 ± 10.3 | 23.7 ± 5.9 * | 81.6 ± 20.4 | 94.7 ± 26.5 | ||||||
| PHE | 120 ± 30 | 230 ± 50 * | 90 ± 30 | 130 ± 40 | ||||||
| Incompatible Interaction | Compatible Interaction | |||
|---|---|---|---|---|
| cntr | +MifieldV | cntr | +MifieldV | |
| NAPDH Ox | 15.0 ± 1.7 | 19.4 ± 2.7 * | 13.1 ± 1.7 | 9.5 ± 1.7 * |
| SOD | 6.7 ± 1.4 | 15.5 ± 1.9 * | 8.6 ± 1.4 | 11.2 ± 1.1 |
| GUA isoPEX | 1.4 ± 0.2 | 1.7 ± 0.3 * | 2.7 ± 0.5 | 1.9 ± 1.0 * |
| SYR isoPEX | 9.5 ± 1.6 | 13.6 ± 2.0 * | 10.4 ± 1.9 | 8.7 ± 1.3 |
| PPD-PC isoPEX | 8.2 ± 1.2 | 9.8 ± 2.1 | 9.9 ± 1.5 | 8.4 ± 1.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molinari, S.; Leonetti, P. Inhibition of ROS-Scavenging Enzyme System Is a Key Event in Tomato Genetic Resistance against Root-Knot Nematodes. Int. J. Mol. Sci. 2023, 24, 7324. https://doi.org/10.3390/ijms24087324
Molinari S, Leonetti P. Inhibition of ROS-Scavenging Enzyme System Is a Key Event in Tomato Genetic Resistance against Root-Knot Nematodes. International Journal of Molecular Sciences. 2023; 24(8):7324. https://doi.org/10.3390/ijms24087324
Chicago/Turabian StyleMolinari, Sergio, and Paola Leonetti. 2023. "Inhibition of ROS-Scavenging Enzyme System Is a Key Event in Tomato Genetic Resistance against Root-Knot Nematodes" International Journal of Molecular Sciences 24, no. 8: 7324. https://doi.org/10.3390/ijms24087324