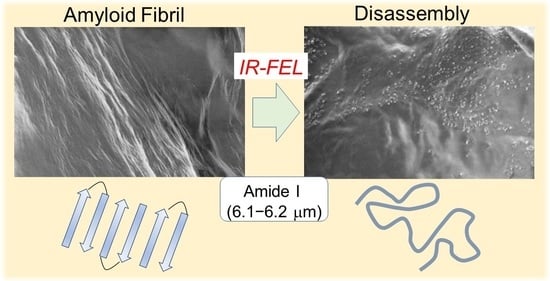
Disassembly of Amyloid Fibril with Infrared Free Electron Laser
Abstract
:1. Introduction
2. Infrared-Laser Induced Dissociation of GNNQQNY Peptide Fibril
2.1. Experimental Results
2.2. Simulation Results
3. Disassembly of β2-Microglobulin Peptide Fibril
3.1. Experimental Results
3.2. Simulation Results
4. Future Application of IR-FEL in Amyloid Research
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blokhuis, A.M.; Groen, E.J.N.; Koppers, M.; van den Berg, L.H.; Pasterkamp, R.J. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013, 125, 777–794. [Google Scholar] [CrossRef]
- Jin, W.; Xing, Z.; Song, Y.; Huang, C.; Xu, X.; Ghose, S.; Li, Z.J. Protein aggregation and mitigation strategy in low pH viral inactivation for monoclonal antibody purification. mAbs 2019, 11, 1479–1491. [Google Scholar] [CrossRef]
- Simpson, L.W.; Good, T.A.; Leach, J.B. Protein folding and assembly in confined environments: Implications for protein aggregation in hydrogels and tissues. Biotechnol. Adv. 2020, 42, 107573. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Xu, Y. Towards modeling of amyloid fibril structures. Front. Biosci. 2008, 13, 4039–4050. [Google Scholar] [CrossRef]
- Marijan, D.; Tse, R.; Elliott, K.; Chandhok, S.; Luo, M.; Lacroix, E.; Audas, T.E. Stress-specific aggregation of proteins in the amyloid bodies. FEBS Lett. 2019, 593, 3162–3172. [Google Scholar] [CrossRef] [PubMed]
- Qiang, W.; Yau, W.M.; Lu, J.X.; Collinge, J.; Tycko, R. Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature 2017, 541, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Benson, M.D.; Buxbaum, J.N.; Eisenberg, D.S.; Merlini, G.; Saraiva, M.J.M.; Sekijima, Y.; Sipe, J.D.; Westermark, P. Amyloid nomenclature 2018: Recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid 2018, 25, 215–219. [Google Scholar] [CrossRef]
- Stroud, J.C.; Liu, C.; Teng, P.K.; Eisenberg, D. Toxic fibrillar oligomers of amyloid-β have cross-β structure. Proc. Natl. Acad. Sci. USA 2012, 109, 7717–7722. [Google Scholar] [CrossRef]
- Benseny-Cases, N.; Cócera, M.; Cladera, J. Conversion of non-fibrillar beta-sheet oligomers into amyloid fibrils in Alzheimer’s disease amyloid peptide aggregation. Biochem. Biophys. Res. Commun. 2007, 361, 916–921. [Google Scholar] [CrossRef]
- Zhu, H.L.; Meng, S.R.; Fan, J.B.; Chen, J.; Liang, Y. Fibrillization of Human Tau Is Accelerated by Exposure to Lead via Interaction with His-330 and His-362. PLoS ONE 2011, 6, e25020. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.H.; Miles, T.F.; Sheehan, M.; Alfieri, K.N.; Kokona, B.; Fairman, R. Polyglutamine fibrils are formed using a simple designed β-hairpin model. Proteins 2010, 78, 1971–1979. [Google Scholar] [CrossRef]
- Zandomeneghi, G.; Krebs, M.R.H.; McCammon, M.G.; Fändrich, M. FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci. 2004, 13, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Daus, M.L.; Wagenführ, K.; Thomzig, A.; Boerner, S.; Hermann, P.; Hermelink, A.; Beekes, M.; Lasch, P. Infrared Microspectroscopy Detects Protein Misfolding Cyclic Amplification (PMCA)-induced Conformational Alterations in Hamster Scrapie Progeny Seeds. J. Biol. Chem. 2013, 288, 35068–35080. [Google Scholar] [CrossRef]
- Botelho, H.M.; Leal, S.S.; Cardoso, I.; Yanamandra, K.; Morozova-Roche, L.A.; Fritz, G.; Gomes, C.M. S100A6 amyloid fibril formation is calcium-modulated and enhances superoxide dismutase-1 (SOD1) aggregation. J. Biol. Chem. 2012, 287, 42233–42242. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.P.; Fink, A.L.; Uversky, V.N. Structural Characteristics of the α-Synuclein Oligomers Stabilized by the Flavonoid Baicalein. J. Mol. Biol. 2008, 383, 214–223. [Google Scholar] [CrossRef]
- Frare, E.; Mossuto, M.F.; Polverino de Laureto, P.; Dumoulin, M.; Dobson, C.M.; Fontana, A. Identification of the Core Structure of Lysozyme Amyloid Fibrils by Proteolysis. J. Mol. Biol. 2006, 361, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Reches, M.; Porat, Y.; Gazit, E. Amyloid Fibril Formation by Pentapeptide and Tetrapeptide Fragments of Human Calcitonin. J. Biol. Chem. 2002, 277, 35475–35480. [Google Scholar] [CrossRef]
- Ahmad, A.; Uversky, V.N.; Hong, D.; Fink, A.L. Early Events in the Fibrillation of Monomeric Insulin. J. Biol. Chem. 2005, 280, 42669–42675. [Google Scholar] [CrossRef]
- Lu, M.; Hiramatsu, H.; Goto, Y.; Kitagawa, T. Structure of Interacting Segments in the Growing Amyloid Fibril of β2-Microglobulin Probed with IR Spectroscopy. J. Mol. Biol. 2006, 362, 355–364. [Google Scholar] [CrossRef]
- Fändrich, M.; Forge, V.; Buder, K.; Kittler, M.; Dobson, C.M.; Diekmann, S. Myoglobin forms amyloid fibrils by association of unfolded polypeptide segments. Proc. Natl. Acad. Sci. USA 2003, 100, 15463–15468. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, T.; Fujioka, J.; Imai, T.; Tsukiyama, K. Effect of Mid-infrared Free-Electron Laser Irradiation on Refolding of Amyloid-Like Fibrils of Lysozyme into Native Form. Protein J. 2012, 31, 710–716. [Google Scholar] [CrossRef]
- Kawasaki, T.; Fujioka, J.; Imai, T.; Torigoe, K.; Tsukiyama, K. Mid-infrared free-electron laser tuned to the amide I band for converting insoluble amyloid-like protein fibrils into the soluble monomeric form. Lasers Med. Sci. 2014, 29, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Newberry, R.W.; Raines, R.T. A prevalent intraresidue hydrogen bond stabilizes proteins. Nat. Chem. Biol. 2016, 12, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.W.; Knowles, T.P.; Waudby, C.A.; Vendruscolo, M.; Dobson, C.M. Inversion of the balance between hydrophobic and hydrogen bonding interactions in protein folding and aggregation. PLoS Comput. Biol. 2011, 7, e1002169. [Google Scholar] [CrossRef] [PubMed]
- Close, W.; Neumann, M.; Schmidt, A.; Hora, M.; Annamalai, K.; Schmidt, M.; Reif, B.; Schmidt, V.; Grigorieff, N.; Fändrich, M. Physical basis of amyloid fibril polymorphism. Nat. Commun. 2018, 9, 699. [Google Scholar] [CrossRef]
- Fändrich, M.; Meinhardt, J.; Grigorieff, N. Structural polymorphism of Alzheimer Abeta and other amyloid fibrils. Prion 2009, 3, 89–93. [Google Scholar] [CrossRef]
- Tycko, R.; Wickner, R.B. Molecular structures of amyloid and prion fibrils: Consensus versus controversy. Acc. Chem. Res. 2013, 46, 1487–1496. [Google Scholar] [CrossRef]
- Grosse, E. THz radiation from free electron lasers and its potential for cell and tissue studies. Phys. Med. Biol. 2002, 47, 3755–3760. [Google Scholar] [CrossRef]
- Knippels, G.M.H.; van de Pol, M.J.; Pellemans, H.P.M.; Planken, P.C.M.; van der Meer, A.F.G. Two-color facility based on a broadly tunable infrared free-electron laser and a subpicosecond-synchronized 10-fs-Ti:sapphire laser. Opt. Lett. 1998, 23, 1754–1756. [Google Scholar] [CrossRef]
- Glotin, F.; Chaput, R.; Jaroszynski, D.; Prazeres, R.; Ortega, J.-M. Infrared subpicosecond laser pulses with a free-electron laser. Phys. Rev. Lett. 1993, 71, 2587–2590. [Google Scholar] [CrossRef]
- Madey, J.M.J. Stimulated Emission of Bremsstrahlung in a Periodic Magnetic Field. J. Appl. Phys. 1971, 42, 1906–1913. [Google Scholar] [CrossRef]
- Edwards, G.; Logan, R.; Copeland, M.; Reinisch, L.; Davidson, J.; Johnson, B.; Maciunas, R.; Mendenhall, M.; Ossoff, R.; Tribble, J.; et al. Tissue ablation by a free-electron laser tuned to the amide II band. Nature 1994, 371, 416–419. [Google Scholar] [CrossRef]
- Vogel, A.; Venugopalan, V. Mechanisms of Pulsed Laser Ablation of Biological Tissues. Chem. Rev. 2003, 103, 577–644. [Google Scholar] [CrossRef]
- Ovelmen-Levitt, J.; Straub, K.D.; Hauger, S.; Szarmes, E.; Madey, J.; Pearlstein, R.D.; Nashold, B.S., Jr. Brain ablation in the rat cerebral cortex using a tunable-free electron laser. Lasers Surg. Med. 2003, 33, 81–92. [Google Scholar] [CrossRef]
- Wells, J.; Kao, C.; Jansen, E.D.; Konrad, P.; Mahadevan-Jansen, A. Application of infrared light for in vivo neural stimulation. J. Biomed. Opt. 2005, 10, 064003. [Google Scholar] [CrossRef]
- Toyama, T.; Fujioka, J.; Watanabe, K.; Yoshida, A.; Sakuma, T.; Inaba, K.; Imai, T.; Nakajima, T.; Tsukiyama, K.; Hamada, N.; et al. Investigation of bactericidal effect of a mid-infrared free electron laser on Escherichia coli. Sci. Rep. 2022, 12, 18111. [Google Scholar] [CrossRef]
- Onami, Y.; Kawasaki, T.; Aizawa, H.; Haraguchi, T.; Akitsu, T.; Tsukiyama, K.; Palafox, M.A. Degradation of Human Serum Albumin by Infrared Free Electron Laser Enhanced by Inclusion of a Salen-Type Schiff Base Zn (II) Complex. Int. J. Mol. Sci. 2020, 21, 874. [Google Scholar] [CrossRef] [PubMed]
- Onami, Y.; Koya, R.; Kawasaki, T.; Aizawa, H.; Nakagame, R.; Miyagawa, Y.; Haraguchi, T.; Akitsu, T.; Tsukiyama, K.; Palafox, M.A. Investigation by DFT Methods of the Damage of Human Serum Albumin Including Amino Acid Derivative Schiff Base Zn(II) Complexes by IR-FEL Irradiation. Int. J. Mol. Sci. 2019, 20, 2846. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, D.E.; Morais, C.L.; Lima, K.M.; Trevisan, J.; Siggel-King, M.R.; Craig, T.; Ingham, J.; Martin, D.S.; Heys, K.; Kyrgiou, M.; et al. An imaging dataset of cervical cells using scanning near-field optical microscopy coupled to an infrared free electron laser. Sci. Data 2017, 4, 170084. [Google Scholar] [CrossRef]
- Feres, F.H.; Mayer, R.A.; Wehmeier, L.; Maia, F.C.B.; Viana, E.R.; Malachias, A.; Bechtel, H.A.; Klopf, J.M.; Eng, L.M.; Kehr, S.C.; et al. Sub-diffractional cavity modes of terahertz hyperbolic phonon polaritons in tin oxide. Nat. Commun. 2021, 12, 1995. [Google Scholar] [CrossRef] [PubMed]
- Irizawa, A.; Fujimoto, M.; Kawase, K.; Kato, R.; Fujiwara, H.; Higashiya, A.; Macis, S.; Tomarchio, L.; Lupi, S.; Marcelli, A.; et al. Spatially Resolved Spectral Imaging by A THz-FEL. Condens. Matter 2020, 5, 38. [Google Scholar] [CrossRef]
- Andersson, Å.; Poline, M.; Kodambattil, M.; Rebrov, O.; Loire, E.; Maître, P.; Zhaunerchyk, V. Structure of Proton-Bound Methionine and Tryptophan Dimers in the Gas Phase Investigated with IRMPD Spectroscopy and Quantum Chemical Calculations. J. Phys. Chem. A 2020, 124, 2408–2415. [Google Scholar] [CrossRef]
- Austin, R.H.; Xie, A.; van der Meer, L.; Redlich, B.; Lindgård, P.-A.; Frauenfelder, H.; Fu, D. Picosecond Thermometer in the Amide I Band of Myoglobin. Phys. Rev. Lett. 2005, 94, 128101. [Google Scholar] [CrossRef]
- Elferink, H.; Severijnen, M.E.; Martens, J.; Mensink, R.A.; Berden, G.; Oomens, J.; Rutjes, F.P.J.T.; Rijs, A.M.; Boltje, T.J. Direct Experimental Characterization of Glycosyl Cations by Infrared Ion Spectroscopy. J. Am. Chem. Soc. 2018, 140, 6034–6038. [Google Scholar] [CrossRef]
- Matsubara, M.; Osada, F.; Nakajima, M.; Imai, T.; Nishimura, K.; Oyama, T.; Tsukiyama, K. Isomerization and dissociation of 2,3-dihydrofuran (2,3-DHF) induced by infrared free electron laser. J. Photochem. Photobiol. A 2016, 322, 53–59. [Google Scholar] [CrossRef]
- Munshi, M.U.; Martens, J.; Berden, G.; Oomens, J. Protoisomerization of Indigo and Isoindigo Dyes Confirmed by Gas-Phase Infrared Ion Spectroscopy. J. Phys. Chem. A 2019, 123, 8226–8233. [Google Scholar] [CrossRef]
- Zavalin, A.; Hachey, D.L.; Sundaramoorthy, M.; Banerjee, S.; Morgan, S.; Feldman, L.; Tolk, N.; Piston, D.W. Kinetics of a Collagen-Like Polypeptide Fragmentation after Mid-IR Free-Electron Laser Ablation. Biophys. J. 2008, 95, 1371–1381. [Google Scholar] [CrossRef]
- Kawasaki, T.; Sakai, T.; Zen, H.; Sumitomo, Y.; Nogami, K.; Hayakawa, K.; Yaji, T.; Ohta, T.; Tsukiyama, K.; Hayakawa, Y. Cellulose Degradation by Infrared Free Electron Laser. Energy Fuels 2020, 34, 9064–9068. [Google Scholar] [CrossRef]
- Kawasaki, T.; Zen, H.; Sakai, T.; Sumitomo, Y.; Nogami, K.; Hayakawa, K.; Yaji, T.; Ohta, T.; Nagata, T.; Hayakawa, Y. Degradation of Lignin by Infrared Free Electron Laser. Polymers 2022, 14, 2401. [Google Scholar] [CrossRef] [PubMed]
- Dienst, A.; Casandruc, E.; Fausti, D.; Zhang, L.; Eckstein, M.; Hoffmann, M.; Khanna, V.; Dean, N.; Gensch, M.; Winnerl, S.; et al. Optical excitation of Josephson plasma solitons in a cuprate superconductor. Nat. Mater. 2013, 12, 535–541. [Google Scholar] [CrossRef]
- Sei, N.; Sakai, T.; Hayakawa, Y.; Sumitomo, Y.; Nogami, K.; Tanaka, T.; Hayakawa, K. Observation of terahertz coherent edge radiation amplified by infrared free-electron laser oscillations. Sci. Rep. 2021, 11, 3433. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Imai, T.; Tsukiyama, K. Use of a Mid-Infrared Free-Electron Laser (MIR-FEL) for Dissociation of the Amyloid Fibril Aggregates of a Peptide. J. Anal. Sci. Methods Instrum. 2014, 4, 9–18. [Google Scholar] [CrossRef]
- Kawasaki, T.; Yaji, T.; Imai, T.; Ohta, T.; Tsukiyama, K. Synchrotron-Infrared Microscopy Analysis of Amyloid Fibrils Irradiated by Mid-Infrared Free-Electron Laser. Am. J. Anal. Chem. 2014, 5, 384–394. [Google Scholar] [CrossRef]
- Kawasaki, T.; Ohori, G.; Chiba, T.; Tsukiyama, K.; Nakamura, K. Picosecond pulsed infrared laser tuned to amide I band dissociates polyglutamine fibrils in cells. Lasers Med. Sci. 2016, 31, 1425–1431. [Google Scholar] [CrossRef]
- Kawasaki, T.; Yaji, T.; Ohta, T.; Tsukiyama, K. Application of mid-infrared free-electron laser tuned to amide bands for dissociation of aggregate structure of protein. J. Synchrotron Rad. 2016, 23, 152–157. [Google Scholar] [CrossRef]
- Kawasaki, T.; Yaji, T.; Ohta, T.; Tsukiyama, K.; Nakamura, K. Dissociation of β-Sheet Stacking of Amyloid β Fibrils by Irradiation of Intense, Short-Pulsed Mid-infrared Laser. Cell. Mol. Neurobiol. 2018, 38, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Tsukiyama, K.; Irizawa, A. Dissolution of a fibrous peptide by terahertz free electron laser. Sci. Rep. 2019, 9, 10636. [Google Scholar] [CrossRef]
- Kawasaki, T.; Izumi, Y.; Ohori, G.; Kitahara, H.; Furuya, T.; Yamamoto, K.; Matsuo, K.; Tani, M.; Tsukiyama, K. Study on Irradiation Effect of Mid-Infrared Free Electron Laser on Hen Egg-White Lysozyme by Using Terahertz-Time Domain Spectroscopy and Synchrotron-Radiation Vacuum-Ultraviolet Circular-Dichroism Spectroscopy. J. Infrared Milli. Terahz Waves 2019, 40, 998–1009. [Google Scholar] [CrossRef]
- Kawasaki, T.; Man, V.H.; Sugimoto, Y.; Sugiyama, N.; Yamamoto, H.; Tsukiyama, K.; Wang, J.; Derreumaux, P.; Nguyen, P.H. Infrared Laser-Induced Amyloid Fibril Dissociation: A Joint Experimental/Theoretical Study on the GNNQQNY Peptide. J. Phys. Chem. B 2020, 124, 6266–6277. [Google Scholar] [CrossRef]
- Jindo, M.; Nakamura, K.; Okumura, H.; Tsukiyama, K.; Kawasaki, T. Application study of infrared free-electron lasers towards the development of amyloidosis therapy. J. Synchrotron Rad. 2022, 29, 1133–1140. [Google Scholar] [CrossRef]
- Kawasaki, T.; Yamaguchi, Y.; Kitahara, H.; Irizawa, A.; Tani, M. Exploring Biomolecular Self-Assembly with Far-Infrared Radiation. Biomolecules 2022, 12, 1326. [Google Scholar] [CrossRef]
- Man, V.H.; Derreumaux, P.; Li, M.S.; Roland, C.; Sagui, C.; Nguyen, P.H. Picosecond dissociation of amyloid fibrils with infrared laser: A nonequilibrium simulation study. J. Chem. Phys. 2015, 143, 155101. [Google Scholar] [CrossRef]
- Man, V.; Truong, P.; Derreumaux, P.; Li, M.; Roland, C.; Sagui, C.; Nguyen, P. Picosecond melting of peptide nanotube with infrared laser: A nonequilibrium study. Phys. Chem. Chem. Phys. 2015, 17, 27275–27280. [Google Scholar] [CrossRef]
- Man, V.H.; Wang, J.; Derreumaux, P.; Nguyen, P.H. Nonequilibrium molecular dynamics simulations of infrared laser-induced dissociation of a tetrameric Aβ42 β-barrel in a neuronal membrane model. Chem. Phys. Lipids 2021, 234, 105030. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Derreumaux, P. Computer Simulations Aimed at Exploring Protein Aggregation and Dissociation. In Computer Simulations of Aggregation of Proteins and Peptides; Li, M.S., Kloczkowski, A., Cieplak, M., Kouza, M., Eds.; Humana: New York, NY, USA, 2022; Volume 2340, pp. 175–196. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Ramamoorthy, A.; Sahoo, B.R.; Zheng, J.; Faller, P.; Straub, J.E.; Dominguez, L.; Shea, J.-E.; Dokholyan, N.V.; De Simone, A.; et al. Amyloid Oligomers: A Joint Experimental/Computational Perspective on Alzheimer’s Disease, Parkinson’s Disease, Type II Diabetes, and Amyotrophic Lateral Sclerosis. Chem. Rev. 2021, 121, 2545–2647. [Google Scholar] [CrossRef]
- Hutson, M.S.; Hauger, S.A.; Edwards, G. Thermal diffusion and chemical kinetics in laminar biomaterial due to heating by a free-electron laser. Phys. Rev. E 2002, 65, 061906. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, A.J.; Knowles, T.P.J.; Tartaglia, G.G.; Fitzpatrick, A.W.; Devlin, G.L.; Shammas, S.L.; Waudby, C.A.; Mossuto, M.F.; Meehan, S.; Gras, S.L.; et al. Metastability of Native Proteins and the Phenomenon of Amyloid Formation. J. Am. Chem. Soc. 2011, 133, 14160–14163. [Google Scholar] [CrossRef]
- Zhang, Y.; Viet, M.H.; Christopher, R.; Sagui, C. Amyloid Properties of Asparagine and Glutamine in Prion-like Proteins. ACS Chem. Neurosci. 2016, 7, 576–587. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.C.; Prusiner, S.B. Experimental Models of Inherited PrP Prion Diseases. Cold Spring Harb. Perspect. Med. 2017, 7, a027151. [Google Scholar] [CrossRef]
- Balbirnie, M.; Grothe, R.; Eisenberg, D.S. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proc. Natl. Acad. Sci. USA 2001, 98, 2375–2380. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.; Sawaya, M.R.; Balbirnie, M.; Madsen, A.Ø.; Riekel, C.; Grothe, R.; Eisenberg, D. Structure of the cross-β spine of amyloid-like fibrils. Nature 2005, 435, 773–778. [Google Scholar] [CrossRef]
- Nasica-Labouze, J.; Meli, M.; Derreumaux, P.; Colombo, G.; Mousseau, N. A Multiscale Approach to Characterize the Early Aggregation Steps of the Amyloid-Forming Peptide GNNQQNY from the Yeast Prion Sup-35. PLoS Comput. Biol. 2011, 7, e1002051. [Google Scholar] [CrossRef]
- Kawasaki, T.; Sato, A.; Tominaga, Y.; Suzuki, Y.; Oyama, T.; Tadokoro, M.; Tsukiyama, K.; Nokihara, K.; Zen, H. Photo-Modification of Melanin by a Mid-Infrared Free-Electron Laser. Photochem. Photobiol. 2019, 95, 946–950. [Google Scholar] [CrossRef] [PubMed]
- Scarpioni, R.; Ricardi, M.; Albertazzi, V.; De Amicis, S.; Rastelli, F.; Zerbini, L. Dialysis-related amyloidosis: Challenges and solutions. Int. J. Nephrol. Renovasc. Dis. 2016, 9, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Nishino, M.; Sugita, Y.; Yoda, T.; Okamoto, Y. Structures of a peptide fragment of β2-microglobulin studied by replica-exchange molecular dynamics simulations—Towards the understanding of the mechanism of amyloid formation. FEBS Lett. 2005, 579, 5425–5429. [Google Scholar] [CrossRef]
- Okumura, H.; Itoh, S.G.; Nakamura, K.; Kawasaki, T. Role of Water Molecules and Helix Structure Stabilization in the Laser-Induced Disruption of Amyloid Fibrils Observed by Nonequilibrium Molecular Dynamics Simulations. J. Phys. Chem. B 2021, 125, 4964–4976. [Google Scholar] [CrossRef] [PubMed]
- Lambracht-Washington, D.; Rosenberg, R.N. Anti-amyloid beta to tau—Based immunization: Developments in immunotherapy for Alzheimer disease. Immunotargets Ther. 2013, 2, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.-F.; Xu, T.-H.; Yan, Y.; Zhou, Y.-R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid beta: Structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef] [PubMed]
- Long, J.M.; Holtzman, D.M. Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 2019, 179, 312–339. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Nguyen, P.H.; Ngo, S.T.; Derreumaux, P. Effect of Cholesterol Molecules on Aβ1-42 Wild-Type and Mutants Trimers. Molecules 2022, 27, 1395. [Google Scholar] [CrossRef]
- Ngo, S.T.; Nguyen, P.H.; Derreumaux, P. Cholesterol Molecules Alter the Energy Landscape of Small Aβ1-42 Oligomers. J. Phys. Chem. B 2021, 125, 2299–2307. [Google Scholar] [CrossRef] [PubMed]
- Nasica-Labouze, J.; Nguyen, P.H.; Sterpone, F.; Berthoumieu, O.; Buchete, N.-V.; Coté, S.; De Simone, A.; Doig, A.J.; Faller, P.; Garcia, A.; et al. Amyloid β Protein and Alzheimer’s Disease: When Computer Simulations Complement Experimental Studies. Chem. Rev. 2015, 115, 3518–3563. [Google Scholar] [CrossRef] [PubMed]
- Habchi, J.; Chia, S.; Galvagnion, C.; Michaels, T.C.T.; Bellaiche, M.M.J.; Ruggeri, F.S.; Sanguanini, M.; Idini, I.; Kumita, J.R.; Sparr, E.; et al. Cholesterol catalyses Aβ42 aggregation through a heterogeneous nucleation pathway in the presence of lipid membranes. Nat. Chem. 2018, 10, 673–683. [Google Scholar] [CrossRef]
- Awazu, K.; Nagai, A.; Aizawa, K. Selective removal of cholesterol esters in an arteriosclerotic region of blood vessels with a free-electron laser. Lasers Surg. Med. 1998, 23, 233–237. [Google Scholar] [CrossRef]
- Chung, H.; Dai, T.; Sharma, S.K.; Huang, Y.Y.; Carroll, J.D.; Hamblin, M.R. The Nuts and Bolts of Low-level Laser (Light) Therapy. Ann. Biomed. Eng. 2012, 40, 516–533. [Google Scholar] [CrossRef]
- Farfara, D.; Tuby, H.; Trudler, D.; Doron-Mandel, E.; Maltz, L.; Vassar, R.J.; Frenkel, D.; Oron, U. Low-Level Laser Therapy Ameliorates Disease Progression in a Mouse Model of Alzheimer’s Disease. J. Mol. Neurosci. 2015, 55, 430–436. [Google Scholar] [CrossRef]
- Jeong, Y.J.; Kang, G.-Y.; Kwon, J.H.; Choi, H.-D.; Pack, J.-K.; Kim, N.; Lee, Y.-S.; Lee, H.-J. 1950 MHz Electromagnetic Fields Ameliorate Aβ Pathology in Alzheimer’s Disease Mice. Curr. Alzheimer Res. 2015, 12, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.R.; Liu, R.T. The Toxicity and Polymorphism of β-Amyloid Oligomers. Int. J. Mol. Sci. 2020, 21, 4477. [Google Scholar] [CrossRef]
- Viola, K.L.; Klein, W.L. Amyloid β oligomers in Alzheimer’s disease pathogenesis, treatment, and diagnosis. Acta Neuropathol. 2015, 129, 183–206. [Google Scholar] [CrossRef]
- Candelise, N.; Scaricamazza, S.; Salvatori, I.; Ferri, A.; Valle, C.; Manganelli, V.; Garofalo, T.; Sorice, M.; Misasi, R. Protein Aggregation Landscape in Neurodegenerative Diseases: Clinical Relevance and Future Applications. Int. J. Mol. Sci. 2021, 22, 6016. [Google Scholar] [CrossRef]
- Fowler, D.M.; Koulov, A.V.; Balch, W.E.; Kelly, J.W. Functional amyloid—From bacteria to humans. Trends Biochem. Sci. 2007, 32, 217–224. [Google Scholar] [CrossRef]
- Hong, S.; Sunwoo, J.H.; Kim, J.S.; Tchah, H.; Hwang, C. Conjugation of carboxymethyl cellulose and dopamine for cell sheet harvesting. Biomater. Sci. 2019, 7, 139–148. [Google Scholar] [CrossRef]
- Qin, C.; Yao, M.; Liu, Y.; Yang, Y.; Zong, Y.; Zhao, H. MFC/NFC-Based Foam/Aerogel for Production of Porous Materials: Preparation, Properties and Applications. Materials 2020, 13, 5568. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.S.; Das, S.; Ghosh, D.; Maji, S.K. Influence of retinoic acid on mesenchymal stem cell differentiation in amyloid hydrogels. Data Brief 2015, 5, 954–958. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Silva, T.L.; Schneider, J.P. From structure to application: Progress and opportunities in peptide materials development. Curr. Opin. Chem. Biol. 2021, 64, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Kokotidou, C.; Jonnalagadda, S.V.R.; Orr, A.A.; Vrentzos, G.; Kretsovali, A.; Tamamis, P.; Mitraki, A.A. Designer Amyloid Cell-Penetrating Peptides for Potential Use as Gene Transfer Vehicles. Biomolecules 2019, 10, 7. [Google Scholar] [CrossRef]
- Dai, B.; Li, D.; Xi, W.; Luo, F.; Zhang, X.; Zou, M.; Cao, M.; Hu, J.; Wang, W.; Wei, G.; et al. Tunable assembly of amyloid-forming peptides into nanosheets as a retrovirus carrier. Proc. Natl. Acad. Sci. USA 2015, 112, 2996–3001. [Google Scholar] [CrossRef]
- Domin, D.; Man, V.H.; Van-Oanh, N.-T.; Wang, J.; Kawasaki, T.; Derreumaux, P.; Nguyen, P.H. Breaking down cellulose fibrils with a mid-infrared laser. Cellulose 2018, 25, 5553–5568. [Google Scholar] [CrossRef]












| Amyloidosis-Related Proteins | Amide I (cm−1) | Ref. |
|---|---|---|
| Aβ1-40/Aβ1-42 (Alzheimer’s disease) | 1630/1626 | [8,9] |
| Tau (Frontotemporal dementia) | 1630 | [10] |
| Polyglutamine (Huntington’s disease) | 1625 | [11] |
| Transthyretin (Amyloidotic polyneuropathy) | 1615 | [12] |
| Prion protein (Creutzfeldt-Jakob disease) | 1626/1635 | [13] |
| S100A6 (Amyotrophic lateral sclerosis) | 1625 | [14] |
| α-Synuclein (Parkinson’s disease) | 1630 | [15] |
| Lysozyme (Hereditary amyloidosis) | 1614 | [16] |
| Calcitonin (Thyroid medullary carcinoma) | 1639 | [17] |
| Insulin (Subcutaneous edema) | 1628 | [18] |
| β2-Microglobulin (Kidney dialysis amyloidosis) | 1629 | [19] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawasaki, T.; Tsukiyama, K.; Nguyen, P.H. Disassembly of Amyloid Fibril with Infrared Free Electron Laser. Int. J. Mol. Sci. 2023, 24, 3686. https://doi.org/10.3390/ijms24043686
Kawasaki T, Tsukiyama K, Nguyen PH. Disassembly of Amyloid Fibril with Infrared Free Electron Laser. International Journal of Molecular Sciences. 2023; 24(4):3686. https://doi.org/10.3390/ijms24043686
Chicago/Turabian StyleKawasaki, Takayasu, Koichi Tsukiyama, and Phuong H. Nguyen. 2023. "Disassembly of Amyloid Fibril with Infrared Free Electron Laser" International Journal of Molecular Sciences 24, no. 4: 3686. https://doi.org/10.3390/ijms24043686