SENP6-Mediated deSUMOylation of VEGFR2 Enhances Its Cell Membrane Transport in Angiogenesis
Abstract
:1. Introduction
2. Results
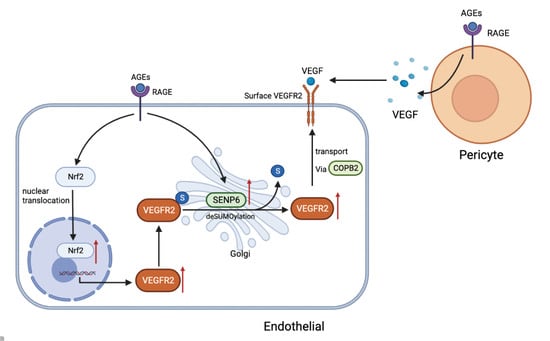
2.1. AGEs Upregulate VEGFR2 Expression by Inducing NF-E2-Related Factor-2 Translocation into the Nucleus
2.2. Inhibiting Nrf2 Entry Decreases AGE−Induced Angiogenesis
2.3. SENP6−Reduced HUVECs Attenuate AGE−Induced Angiogenic Signaling
2.4. VEGFR2 SUMOylation Retains VEGFR2 in the Golgi
2.5. The Vesicular Transport Protein COPB2 Mediates Enhanced Transport of VEGFR2 to Membranes
2.6. AGEs Stimulate Increased Secretion of VEGF by Pericytes and Promote Endothelial Cell Angiogenesis
3. Discussion
4. Materials and Methods
4.1. Culture of HUVECs
4.2. Isolation of Retinal Microvascular Pericytes
4.3. Preparation of AGEs-Modified Bovine Serum Protein
4.4. RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction
4.5. Extraction of Cytoplasmic Protein and Nuclear Protein
4.6. Extraction of Membrane Protein and Cytoplasmic Protein
4.7. Western Blot Analysis
4.8. Co-Immunoprecipitation
4.9. Enzyme-Linked Immunosorbent Assay (ELISA)
4.10. Immunofluorescent Test
4.11. Transfection of siRNA
4.12. Cell Counting Kit-8 (CCK-8)
4.13. Endothelial Cell Transwell Assay
4.14. Scratch Assay for Migration Cells
4.15. EC Tube Formation Assay
4.16. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barrett, E.J.; Liu, Z.; Khamaisi, M.; King, G.L.; Klein, R.; Klein, B.E.K.; Hughes, T.M.; Craft, S.; Freedman, B.I.; Bowden, D.W.; et al. Diabetic microvascular disease: An endocrine society scientific statement. J. Clin. Endocrinol. Metab. 2017, 102, 4343–4410. [Google Scholar] [CrossRef]
- Faselis, C.; Katsimardou, A.; Imprialos, K.; Deligkaris, P.; Kallistratos, M.; Dimitriadis, K. Microvascular Complications of Type 2 Diabetes Mellitus. Curr. Vasc. Pharmacol. 2020, 18, 117–124. [Google Scholar] [CrossRef]
- Tarr, J.M.; Kaul, K.; Wolanska, K.; Kohner, E.M.; Chibber, R. Retinopathy in diabetes. Adv. Exp. Med. Biol. 2012, 771, 88–106. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.Y.; Cheung, C.M.; Larsen, M.; Sharma, S.; Simó, R. Diabetic retinopathy. Nat. Rev. Dis. Primers 2016, 2, 16012. [Google Scholar] [CrossRef] [PubMed]
- Stitt, A.W.; Curtis, T.M.; Chen, M.; Medina, R.J.; McKay, G.J.; Jenkins, A.; Gardiner, T.A.; Lyons, T.J.; Hammes, H.P.; Simó, R.; et al. The progress in understanding and treatment of diabetic retinopathy. Prog. Retin. Eye Res. 2016, 51, 156–186. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P.; Jain, R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature 2011, 473, 298–307. [Google Scholar] [CrossRef] [Green Version]
- Duran, C.L.; Howell, D.W.; Dave, J.M.; Smith, R.L.; Torrie, M.E.; Essner, J.J.; Bayless, K.J. Molecular Regulation of Sprouting Angiogenesis. Compr. Physiol. 2017, 8, 153–235. [Google Scholar] [CrossRef]
- Kim, J.; Kim, Y.H.; Kim, J.; Park, D.Y.; Bae, H.; Lee, D.H.; Kim, K.H.; Hong, S.P.; Jang, S.P.; Kubota, Y.; et al. YAP/TAZ regulates sprouting angiogenesis and vascular barrier maturation. J. Clin. Investig. 2017, 127, 3441–3461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eilken, H.M.; Adams, R.H. Dynamics of endothelial cell behavior in sprouting angiogenesis. Curr. Opin. Cell Biol. 2010, 22, 617–625. [Google Scholar] [CrossRef]
- Cho, H.; Kambhampati, S.P.; Lai, M.J.; Zhou, L.; Lee, G.; Xie, Y.; Hui, Q.; Kannan, R.M.; Duh, E.J. Dendrimer-Triamcinolone Acetonide Reduces Neuroinflammation, Pathological Angiogenesis, and Neuroretinal Dysfunction in Ischemic Retinopathy. Adv. Ther. 2021, 4, 2000181. [Google Scholar] [CrossRef]
- Basta, G.; Schmidt, A.M.; De Caterina, R. Advanced glycation end products and vascular inflammation: Implications for accelerated atherosclerosis in diabetes. Cardiovasc. Res. 2004, 63, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Manigrasso, M.B.; Juranek, J.; Ramasamy, R.; Schmidt, A.M. Unlocking the biology of RAGE in diabetic microvascular complications. Trends Endocrinol. Metab. 2014, 25, 15–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowotny, K.; Jung, T.; Höhn, A.; Weber, D.; Grune, T. Advanced glycation end products and oxidative stress in type 2 diabetes mellitus. Biomolecules 2015, 5, 194–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patche, J.; Girard, D.; Catan, A.; Boyer, F.; Dobi, A.; Planesse, C.; Diotel, N.; Guerin-Dubourg, A.; Baret, P.; Bravo, S.B.; et al. Diabetes-induced hepatic oxidative stress: A new pathogenic role for glycated albumin. Free Radic. Biol. Med. 2017, 102, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Tammela, T.; Enholm, B.; Alitalo, K.; Paavonen, K. The biology of vascular endothelial growth factors. Cardiovasc. Res. 2005, 65, 550–563. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef]
- Ferrara, N. Vascular endothelial growth factor: Basic science and clinical progress. Endocr. Rev. 2004, 25, 581–611. [Google Scholar] [CrossRef]
- Simons, M. An inside view: VEGF receptor trafficking and signaling. Physiology 2012, 27, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Shi, D.S.; Winter, J.M.; Rich, B.E.; Tong, Z.; Sorensen, L.K.; Zhao, H.; Huang, Y.; Tai, Z.; Mleynek, T.M.; et al. Small GTPase ARF6 controls VEGFR2 trafficking and signaling in diabetic retinopathy. J. Clin. Investig. 2017, 127, 4569–4582. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.M.; Yeh, E.T.H. SUMO: From Bench to Bedside. Physiol. Rev. 2020, 100, 1599–1619. [Google Scholar] [CrossRef]
- Rodríguez, J.A. Interplay between nuclear transport and ubiquitin/SUMO modifications in the regulation of cancer-related proteins. Semin. Cancer Biol. 2014, 27, 11–19. [Google Scholar] [CrossRef]
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef]
- Yeh, E.T.; Gong, L.; Kamitani, T. Ubiquitin-like proteins: New wines in new bottles. Gene 2000, 248, 1–14. [Google Scholar] [CrossRef]
- Huang, C.; Han, Y.; Wang, Y.; Sun, X.; Yan, S.; Yeh, E.T.; Chen, Y.; Cang, H.; Li, H.; Shi, G.; et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 2009, 28, 2748–2762. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Teng, X.L.; Zhang, T.; Yu, X.; Ding, R.; Yi, J.; Deng, L.; Wang, Z.; Zou, Q. SENP3 senses oxidative stress to facilitate STING-dependent dendritic cell antitumor function. Mol. Cell 2021, 81, 940–952.e945. [Google Scholar] [CrossRef]
- Di Bacco, A.; Ouyang, J.; Lee, H.Y.; Catic, A.; Ploegh, H.; Gill, G. The SUMO-specific protease SENP5 is required for cell division. Mol. Cell Biol. 2006, 26, 4489–4498. [Google Scholar] [CrossRef] [Green Version]
- Jansen, N.S.; Vertegaal, A.C.O. A Chain of Events: Regulating Target Proteins by SUMO Polymers. Trends Biochem. Sci. 2021, 46, 113–123. [Google Scholar] [CrossRef]
- Lara-Ureña, N.; Jafari, V.; García-Domínguez, M. Cancer-Associated Dysregulation of Sumo Regulators: Proteases and Ligases. Int. J. Mol. Sci. 2022, 23, 8012. [Google Scholar] [CrossRef]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Wei, Y.; Gong, J.; Thimmulappa, R.K.; Kosmider, B.; Biswal, S.; Duh, E.J. Nrf2 acts cell-autonomously in endothelium to regulate tip cell formation and vascular branching. Proc. Natl. Acad. Sci. USA 2013, 110, E3910–E3918. [Google Scholar] [CrossRef]
- Mi, Y.; Yu, M.; Zhang, L.; Sun, C.; Wei, B.; Ding, W.; Zhu, Y.; Tang, J.; Xia, G.; Zhu, L. COPB2 Is Upregulated in Prostate Cancer and Regulates PC-3 Cell Proliferation, Cell Cycle, and Apoptosis. Arch. Med. Res. 2016, 47, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Sun, C.; Zhang, L.; Wan, H.; Zhou, H.; Chen, Y.; Zhu, L.; Xia, G.; Mi, Y. Upregulation of COPB2 Promotes Prostate Cancer Proliferation and Invasion Through the MAPK/TGF-β Signaling Pathway. Front. Oncol. 2022, 12, 865317. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Lei, X.; Zhang, L.; Wan, H.; Pan, H.; Wu, J.; Zou, M.; Zhu, L.; Mi, Y. COPB2: A transport protein with multifaceted roles in cancer development and progression. Clin. Transl. Oncol. 2021, 23, 2195–2205. [Google Scholar] [CrossRef]
- Rezzola, S.; Belleri, M.; Gariano, G.; Ribatti, D.; Costagliola, C.; Semeraro, F.; Presta, M. In vitro and ex vivo retina angiogenesis assays. Angiogenesis 2014, 17, 429–442. [Google Scholar] [CrossRef]
- Li, P.; Chen, D.; Cui, Y.; Zhang, W.; Weng, J.; Yu, L.; Chen, L.; Chen, Z.; Su, H.; Yu, S.; et al. Src Plays an Important Role in AGE−Induced Endothelial Cell Proliferation, Migration, and Tubulogenesis. Front. Physiol. 2018, 9, 765. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Fan, A.; Yuan, Y.; Chen, L.; Guo, X.; Huang, X.; Huang, Q. Role of Moesin in Advanced Glycation End Products-Induced Angiogenesis of Human Umbilical Vein Endothelial Cells. Sci. Rep. 2016, 6, 22749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tezuka, M.; Koyama, N.; Morisaki, N.; Saito, Y.; Yoshida, S.; Araki, N.; Horiuchi, S. Angiogenic effects of advanced glycation end products of the Maillard reaction on cultured human umbilical cord vein endothelial cells. Biochem. Biophys. Res. Commun. 1993, 193, 674–680. [Google Scholar] [CrossRef]
- Devi, M.S.; Sudhakaran, P.R. Differential modulation of angiogenesis by advanced glycation end products. Exp. Biol. Med. 2011, 236, 52–61. [Google Scholar] [CrossRef]
- Xie, Y.; You, S.J.; Zhang, Y.L.; Han, Q.; Cao, Y.J.; Xu, X.S.; Yang, Y.P.; Li, J.; Liu, C.F. Protective role of autophagy in AGE−induced early injury of human vascular endothelial cells. Mol. Med. Rep. 2011, 4, 459–464. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Chang, Y.; Ye, N.; Dai, D.; Chen, Y.; Zhang, N.; Sun, G.; Sun, Y. Advanced Glycation End Products Inhibit the Proliferation of Human Umbilical Vein Endothelial Cells by Inhibiting Cathepsin D. Int. J. Mol. Sci. 2017, 18, 436. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, X.; Yang, S.; Song, X.; Sun, N.; Chen, C.; Zhang, Y.; Yao, D.; Huang, J.; Wang, J.; et al. Kanglexin accelerates diabetic wound healing by promoting angiogenesis via FGFR1/ERK signaling. Biomed. Pharmacother. 2020, 132, 110933. [Google Scholar] [CrossRef]
- Roca, F.; Grossin, N.; Chassagne, P.; Puisieux, F.; Boulanger, E. Glycation: The angiogenic paradox in aging and AGE−related disorders and diseases. Ageing Res. Rev. 2014, 15, 146–160. [Google Scholar] [CrossRef]
- Kaspar, J.W.; Niture, S.K.; Jaiswal, A.K. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 2009, 47, 1304–1309. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ma, W.; Wang, P.Y.; Hurley, P.J.; Bunz, F.; Hwang, P.M. Polo-like kinase 2 activates an antioxidant pathway to promote the survival of cells with mitochondrial dysfunction. Free Radic. Biol. Med. 2014, 73, 270–277. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Lv, H.; Li, J.; Che, Y.; Xu, B.; Tao, Z.; Jiang, W. Roles of Nrf2/HO-1 and HIF-1α/VEGF in lung tissue injury and repair following cerebral ischemia/reperfusion injury. J. Cell Physiol. 2019, 234, 7695–7707. [Google Scholar] [CrossRef]
- Rahimi, N.; Costello, C.E. Emerging roles of post-translational modifications in signal transduction and angiogenesis. Proteomics 2015, 15, 300–309. [Google Scholar] [CrossRef] [Green Version]
- Hickey, C.M.; Wilson, N.R.; Hochstrasser, M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012, 13, 755–766. [Google Scholar] [CrossRef] [Green Version]
- Vertegaal, A.C.O. Signalling mechanisms and cellular functions of SUMO. Nat. Rev. Mol. Cell Biol. 2022, 23, 715–731. [Google Scholar] [CrossRef]
- Zhou, H.J.; Xu, Z.; Wang, Z.; Zhang, H.; Zhuang, Z.W.; Simons, M.; Min, W. SUMOylation of VEGFR2 regulates its intracellular trafficking and pathological angiogenesis. Nat. Commun. 2018, 9, 3303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simons, M.; Gordon, E.; Claesson-Welsh, L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 2016, 17, 611–625. [Google Scholar] [CrossRef]
- Yamada, K.H.; Nakajima, Y.; Geyer, M.; Wary, K.K.; Ushio-Fukai, M.; Komarova, Y.; Malik, A.B. KIF13B regulates angiogenesis through Golgi to plasma membrane trafficking of VEGFR2. J. Cell Sci. 2014, 127, 4518–4530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef] [Green Version]
- Goel, H.L.; Mercurio, A.M. VEGF targets the tumour cell. Nat. Rev. Cancer 2013, 13, 871–882. [Google Scholar] [CrossRef]
- Chang, M.; Nguyen, T.T. Strategy for Treatment of Infected Diabetic Foot Ulcers. Acc. Chem. Res. 2021, 54, 1080–1093. [Google Scholar] [CrossRef]
- Moreau, R. VEGF-induced angiogenesis drives collateral circulation in portal hypertension. J. Hepatol. 2005, 43, 6–8. [Google Scholar] [CrossRef]
- Rattner, A.; Williams, J.; Nathans, J. Roles of HIFs and VEGF in angiogenesis in the retina and brain. J. Clin. Investig. 2019, 129, 3807–3820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossino, M.G.; Lulli, M.; Amato, R.; Cammalleri, M.; Monte, M.D.; Casini, G. Oxidative Stress Induces a VEGF Autocrine Loop in the Retina: Relevance for Diabetic Retinopathy. Cells 2020, 9, 1452. [Google Scholar] [CrossRef] [PubMed]
- Froger, N.; Matonti, F.; Roubeix, C.; Forster, V.; Ivkovic, I.; Brunel, N.; Baudouin, C.; Sahel, J.A.; Picaud, S. VEGF is an autocrine/paracrine neuroprotective factor for injured retinal ganglion neurons. Sci. Rep. 2020, 10, 12409. [Google Scholar] [CrossRef]
- Caporali, A.; Martello, A.; Miscianinov, V.; Maselli, D.; Vono, R.; Spinetti, G. Contribution of pericyte paracrine regulation of the endothelium to angiogenesis. Pharmacol. Ther. 2017, 171, 56–64. [Google Scholar] [CrossRef]
- Eilken, H.M.; Diéguez-Hurtado, R.; Schmidt, I.; Nakayama, M.; Jeong, H.W.; Arf, H.; Adams, S.; Ferrara, N.; Adams, R.H. Pericytes regulate VEGF-induced endothelial sprouting through VEGFR1. Nat. Commun. 2017, 8, 1574. [Google Scholar] [CrossRef]
- Ribatti, D.; Nico, B.; Crivellato, E. The role of pericytes in angiogenesis. Int. J. Dev. Biol. 2011, 55, 261–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.S.; Hu, J.Q.; Liu, X.H.; Chen, L.X.; Chen, H.; Guo, X.H.; Huang, Q.B. Role of Moesin Phosphorylation in Retinal Pericyte Migration and Detachment Induced by Advanced Glycation Endproducts. Front. Endocrinol. 2020, 11, 603450. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cui, Y.; Li, B.; Weng, J.; Wang, W.; Zhang, S.; Huang, X.; Guo, X.; Huang, Q. Advanced glycation end products induce immature angiogenesis in in vivo and ex vivo mouse models. Am. J. Physiol. Heart Circ. Physiol. 2020, 318, H519–H533. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 2015, 72, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, Y.; Ekman, N.; Wu, Z.; Wu, J.; Alitalo, K.; Min, W. Etk/Bmx transactivates vascular endothelial growth factor 2 and recruits phosphatidylinositol 3-kinase to mediate the tumor necrosis factor-induced angiogenic pathway. J. Biol. Chem. 2003, 278, 51267–51276. [Google Scholar] [CrossRef] [PubMed]






| Name | Sequence |
|---|---|
| SENP6 forward | CGGGTGCGGCCATTT |
| SENP6 reverse | GCCGTGGGTTCCCAAGA |
| Gja1 forward | GACTGCGGATCTCCAAAATA |
| Gja1 reverse | CTGTAATTCGCCCAGTTTTG |
| YIPF5 forward | GTAGCAGATGGCAGCATCAT |
| YIPF5 reverse | TGCCAGCCAGTAGCAATGTG |
| CTSC forward | CCCTGGGAGATATGATTAGGAGA |
| CTSC reverse | CAGTCAGTGGTGCAGGTTTG |
| LMAN1 forward | GGTGATCTTTCCATTGGTG |
| LMAN1 reverse | TCCGCTGTATGGTTACTTTG |
| COPB2 forward | CTTCCTGTTCGAGCTGCAAAG |
| COPB2 reverse | CACTCTAATCTGCATGTCATCCG |
| CLTC forward | GCACTGAAAGCTGGGAAAACT |
| CLTC reverse | CTGCAAGGCTAGAATGGCGA |
| Name | Sequence |
|---|---|
| NC siRNA Sense | UUCUCCGAACGUGUCACGUTT |
| NCsiRNAAntisense | ACGUGACACGUUCGGAGAAT |
| Nrf2siRNA Sense | GGAGGCAAGAUAUAGAUCUTT |
| Nrf2siRNAAntisense | AGAUCUAUAUCUUGCCUCCT |
| SENP6siRNA Sense | CCAAGGUGUUGAACGUAUATT |
| SENP6siRNAAntisense | UAUACGUUCAACACCUUGGTT |
| COPB2siRNA Sense | CCCAUUAUGUUAUGCAGAUTT |
| COPB2siRNAAntisense | AUCUGCAUAACAUAAUGGGTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Q.; Chen, Z.; Li, J.; Liu, J.; Zuo, Z.; Lin, B.; Song, K.; Zhou, C.; Lai, H.; Huang, Q.; et al. SENP6-Mediated deSUMOylation of VEGFR2 Enhances Its Cell Membrane Transport in Angiogenesis. Int. J. Mol. Sci. 2023, 24, 2544. https://doi.org/10.3390/ijms24032544
He Q, Chen Z, Li J, Liu J, Zuo Z, Lin B, Song K, Zhou C, Lai H, Huang Q, et al. SENP6-Mediated deSUMOylation of VEGFR2 Enhances Its Cell Membrane Transport in Angiogenesis. International Journal of Molecular Sciences. 2023; 24(3):2544. https://doi.org/10.3390/ijms24032544
Chicago/Turabian StyleHe, Qi, Zhenfeng Chen, Jieyu Li, Jinlian Liu, Zirui Zuo, Bingqi Lin, Ke Song, Chuyu Zhou, Haipeng Lai, Qiaobing Huang, and et al. 2023. "SENP6-Mediated deSUMOylation of VEGFR2 Enhances Its Cell Membrane Transport in Angiogenesis" International Journal of Molecular Sciences 24, no. 3: 2544. https://doi.org/10.3390/ijms24032544