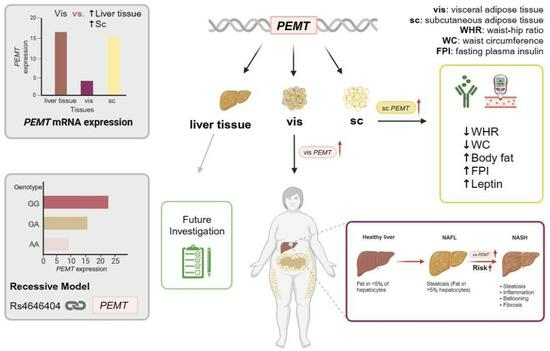
The Role of Phosphatidylethanolamine N-Methyltransferase (PEMT) and Its Waist-Hip-Ratio-Associated Locus rs4646404 in Obesity-Related Metabolic Traits and Liver Disease
Abstract
:1. Introduction
2. Results
2.1. PEMT mRNA Expression in AT Correlates with Parameters of Obesity, Body Fat Distribution, Insulin Sensitivity and Circulating Adipokines
2.2. Association of PEMT Expression with Metabolic Traits and Presence of NAFLD and NASH
2.3. Decreased Lipid Accumulation in Pemt Knockdown in Epididymal Adipocyte Cell Line
2.4. Association of rs4646404 in PEMT with Fat Depot-Specific PEMT mRNA
2.5. rs4646404 and Parameters Related to Obesity, Diabetes and Fat Distribution
3. Discussion
3.1. PEMT mRNA Expression Is Fat Depot Specific and Related to Diabetes Status and NASH
3.2. SNP rs4646404: A Potential Modulator of PEMT Expression
4. Materials and Methods
4.1. Subjects
4.2. PEMT mRNA Expression Analysis in AT and Liver
4.3. Genotyping
4.4. In Vitro Pemt Knockdown in Murine Epidydymal and Inguinal Cell Lines
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Heymsfield, S.B.; Wadden, T.A. Mechanisms, Pathophysiology, and Management of Obesity. N. Engl. J. Med. 2017, 376, 254–266. [Google Scholar] [CrossRef]
- Blüher, M. Adipose Tissue Dysfunction in Obesity. Exp. Clin. Endocrinol. Diabetes 2009, 117, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-Wide Polygenic Scores for Common Diseases Identify Individuals with Risk Equivalent to Monogenic Mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef] [PubMed]
- Khera, A.V.; Chaffin, M.; Wade, K.H.; Zahid, S.; Brancale, J.; Xia, R.; Distefano, M.; Senol-Cosar, O.; Haas, M.E.; Bick, A.; et al. Polygenic Prediction of Weight and Obesity Trajectories from Birth to Adulthood. Cell 2019, 177, 587–596.e9. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Kovacs, P.; Guiu-Jurado, E. Genetics of Obesity in East Asians. Front. Genet. 2020, 11, 1224. [Google Scholar] [CrossRef]
- Sun, C.; Kovacs, P.; Guiu-Jurado, E. Genetics of Body Fat Distribution: Comparative Analyses in Populations with European, Asian and African Ancestries. Genes 2021, 12, 841. [Google Scholar] [CrossRef]
- Shungin, D.; Winkler, T.; Croteau-Chonka, D.C.; Ferreira, T.; Locke, A.E.; Mägi, R.; Strawbridge, R.J.; Pers, T.H.; Fischer, K.; Justice, A.E.; et al. New Genetic Loci Link Adipose and Insulin Biology to Body Fat Distribution. Nature 2015, 518, 187–196. [Google Scholar] [CrossRef]
- Vance, D.E. Physiological Roles of Phosphatidylethanolamine N-Methyltransferase. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2013, 1831, 626–632. [Google Scholar] [CrossRef]
- Li, Z.; Agellon, L.B.; Allen, T.M.; Umeda, M.; Jewell, L.; Mason, A.; Vance, D.E. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006, 3, 321–331. [Google Scholar] [CrossRef]
- Puri, P.; Baillie, R.A.; Wiest, M.M.; Mirshahi, F.; Choudhury, J.; Cheung, O.; Sargeant, C.; Contos, M.J.; Sanyal, A.J. A Lipidomic Analysis of Nonalcoholic Fatty Liver Disease. Hepatology 2007, 46, 1081–1090. [Google Scholar] [CrossRef]
- Piras, I.S.; Raju, A.; Don, J.; Schork, N.J.; Gerhard, G.S.; DiStefano, J.K. Hepatic PEMT Expression Decreases with Increasing NAFLD Severity. Int. J. Mol. Sci. 2022, 23, 9296. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, R.L.; Zhao, Y.; Koonen, D.P.Y.; Sletten, T.; Su, B.; Lingrell, S.; Cao, G.; Peake, D.A.; Kuo, M.S.; Proctor, S.D.; et al. Impaired de Novo Choline Synthesis Explains Why Phosphatidylethanolamine N-Methyltransferase-Deficient Mice Are Protected from Diet-Induced Obesity. J. Biol. Chem. 2010, 285, 22403–22413. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; van der Veen, J.N.; N’Goma, J.C.B.; Nelson, R.C.; Vance, D.E.; Jacobs, R.L. Hepatic PEMT Activity Mediates Liver Health, Weight Gain, and Insulin Resistance. FASEB J. 2019, 33, 10986–10995. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.K.; Langberg, K.A.; Mondal, A.K.; Das, S.K. Phospholipid Biosynthesis Genes and Susceptibility to Obesity: Analysis of Expression and Polymorphisms. PLoS ONE 2013, 8, e65303. [Google Scholar] [CrossRef]
- Presa, N.; Dominguez-Herrera, A.; van der Veen, J.N.; Vance, D.E.; Gómez-Muñoz, A. Implication of Phosphatidylethanolamine N-Methyltransferase in Adipocyte Differentiation. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165853. [Google Scholar] [CrossRef]
- Hörl, G.; Wagner, A.; Cole, L.K.; Malli, R.; Reicher, H.; Kotzbeck, P.; Köfeler, H.; Höfler, G.; Frank, S.; Bogner-Strauss, J.G.; et al. Sequential Synthesis and Methylation of Phosphatidylethanolamine Promote Lipid Droplet Biosynthesis and Stability in Tissue Culture and in Vivo. J. Biol. Chem. 2011, 286, 17338–17350. [Google Scholar] [CrossRef]
- Van Der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The Critical Role of Phosphatidylcholine and Phosphatidylethanolamine Metabolism in Health and Disease. Biochim. Biophys. Acta BBA-Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Cole, L.K.; Vance, D.E. A Role for Sp1 in Transcriptional Regulation of Phosphatidylethanolamine N-Methyltransferase in Liver and 3T3-L1 Adipocytes. J. Biol. Chem. 2010, 285, 11880–11891. [Google Scholar] [CrossRef]
- Walker, A.K.; Jacobs, R.L.; Watts, J.L.; Rottiers, V.; Jiang, K.; Finnegan, D.M.; Shioda, T.; Hansen, M.; Yang, F.; Niebergall, L.J.; et al. A Conserved SREBP-1/Phosphatidylcholine Feedback Circuit Regulates Lipogenesis in Metazoans. Cell 2011, 147, 840–852. [Google Scholar] [CrossRef]
- Fu, S.; Yang, L.; Li, P.; Hofmann, O.; Dicker, L.; Hide, W.; Lin, X.; Watkins, S.M.; Ivanov, A.R.; Hotamisligil, G.S. Aberrant Lipid Metabolism Disrupts Calcium Homeostasis Causing Liver Endoplasmic Reticulum Stress in Obesity. Nature 2011, 473, 528–531. [Google Scholar] [CrossRef]
- Martínez-Uña, M.; Varela-Rey, M.; Cano, A.; Fernández-Ares, L.; Beraza, N.; Aurrekoetxea, I.; Martínez-Arranz, I.; García-Rodríguez, J.L.; Buqué, X.; Mestre, D.; et al. Excess S-Adenosylmethionine Reroutes Phosphatidylethanolamine towards Phosphatidylcholine and Triglyceride Synthesis. Hepatology 2013, 58, 1296–1305. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, R.L.; van der Veen, J.N.; Vance, D.E. Finding the Balance: The Role of S-Adenosylmethionine and Phosphatidylcholine Metabolism in Development of Nonalcoholic Fatty Liver Disease. Hepatology 2013, 58, 1207–1209. [Google Scholar] [CrossRef]
- Kmiec, Z. Cooperation of Liver Cells in Health and Disease. Adv. Anat. Embryol. Cell Biol. 2001, 161, II–XIII, 1–151. [Google Scholar] [CrossRef]
- Mold, C. Effect of Membrane Phospholipids on Activation of the Alternative Complement Pathway. J. Immunol. 1989, 143, 1663–1668. [Google Scholar] [CrossRef] [PubMed]
- Justice, A.E.; Winkler, T.W.; Feitosa, M.F.; Graff, M.; Fisher, V.A.; Young, K.; Barata, L.; Deng, X.; Czajkowski, J.; Hadley, D.; et al. Genome-Wide Meta-Analysis of 241,258 Adults Accounting for Smoking Behaviour Identifies Novel Loci for Obesity Traits. Nat. Commun. 2017, 8, 14977. [Google Scholar] [CrossRef]
- Winkler, T.W.; Justice, A.E.; Graff, M.; Barata, L.; Feitosa, M.F.; Chu, S.; Czajkowski, J.; Esko, T.; Fall, T.; Kilpeläinen, T.O.; et al. The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study. PLoS Genet. 2015, 11, e1005378. [Google Scholar] [CrossRef]
- GTEx Portal. Available online: https://gtexportal.org/home/snp/rs4646404 (accessed on 20 October 2023).
- Freedman, M.L.; Monteiro, A.N.A.; Gayther, S.A.; Coetzee, G.A.; Risch, A.; Plass, C.; Casey, G.; De Biasi, M.; Carlson, C.; Duggan, D.; et al. Principles for the Post-GWAS Functional Characterization of Cancer Risk Loci. Nat. Genet. 2011, 43, 513. [Google Scholar] [CrossRef]
- Klöting, N.; Fasshauer, M.; Dietrich, A.; Kovacs, P.; Schön, M.R.; Kern, M.; Stumvoll, M.; Blüher, M. Insulin-Sensitive Obesity. Am. J. Physiol. Endocrinol. Metab. 2010, 299, 506–515. [Google Scholar] [CrossRef]
- Blüher, M.; Unger, R.; Rassoul, F.; Richter, V.; Paschke, R. Relation between Glycaemic Control, Hyperinsulinaemia and Plasma Concentrations of Soluble Adhesion Molecules in Patients with Impaired Glucose Tolerance or Type II Diabetes. Diabetologia 2002, 45, 210–216. [Google Scholar] [CrossRef]
- Association, A.D. Diagnosis and Classification of Diabetes Mellitus. Diabetes Care 2014, 37, S81–S90. [Google Scholar] [CrossRef]
- Lange, U.G.; Moulla, Y.; Schütz, T.; Blüher, M.; Peter, V.; Shang, E.; Dietrich, A. Effectiveness and Tolerability of a Two-Week Hypocaloric Protein-Rich Diet Prior to Obesity Surgery with Two Different Diet Interventions: A Prospective Randomized Trial. Obes. Surg. 2022, 32, 2903. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and Validation of a Histological Scoring System for Nonalcoholic Fatty Liver Disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Mardinoglu, A.; Heiker, J.T.; Gärtner, D.; Björnson, E.; Schön, M.R.; Flehmig, G.; Klöting, N.; Krohn, K.; Fasshauer, M.; Stumvoll, M.; et al. Extensive Weight Loss Reveals Distinct Gene Expression Changes in Human Subcutaneous and Visceral Adipose Tissue OPEN. Sci. Rep. 2015, 5, 14841. [Google Scholar] [CrossRef] [PubMed]
- Gesta, S.; Blüher, M.; Yamamoto, Y.; Norris, A.W.; Berndt, J.; Kralisch, S.; Boucher, J.; Lewis, C.; Kahn, C.R. Evidence for a Role of Developmental Genes in the Origin of Obesity and Body Fat Distribution. Proc. Natl. Acad. Sci. USA 2006, 103, 6676–6681. [Google Scholar] [CrossRef]
- Klein, J.; Fasshauer, M.; Ito, M.; Lowell, B.B.; Benito, M.; Kahn, C.R. Β3-Adrenergic Stimulation Differentially Inhibits Insulin Signaling and Decreases Insulin-Induced Glucose Uptake in Brown Adipocytes. J. Biol. Chem. 1999, 274, 34795–34802. [Google Scholar] [CrossRef]
- Kovsan, J.; Osnis, A.; Maissel, A.; Mazor, L.; Tarnovscki, T.; Hollander, L.; Ovadia, S.; Meier, B.; Klein, J.; Bashan, N.; et al. Depot-Specific Adipocyte Cell Lines Reveal Differential Drug-Induced Responses of White Adipocytes—Relevance for Partial Lipodystrophy. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E315–E322. [Google Scholar] [CrossRef]






| Sc AT PEMT mRNA p-Value adj (Beta; [95% CI]) | Vis AT PEMT mRNA p-Value adj (Beta; [95% CI]) | Liver PEMT mRNA p-Value adj (Beta; [95% CI]) | |
|---|---|---|---|
| Age (years) | 0.5 (0.69; [−1.38 2.77]) | 0.59 (2.48; [−6.85 11.80]) | 0.5 (−0.33; [−1.29 0.64]) |
| Body weight (kg) | 0.09 (2.75; [−0.40 5.90]) | 0.75 (2.2; [−11.96 16.37]) | 0.58 (−0.44; [−2.03 1.16]) |
| Height (m) | 0.21 (0.01; [−0.01 0.03]) | 0.92 (0; [−0.07 0.06]) | 0.86 (0; [−0.007 0.008]) |
| BMI (kg/m²) | 0.26 (0.56; [−0.43 1.55]) | 0.57 (1.26; [−32,230 5.76]) | 0.35 (−0.23; [−0.71 0.25]) |
| Waist circumference (cm) | 0.01 (2.28; [0.58 3.97]) | 0.32 (3.89; [−3.95 11.72]) | 0.25 (−0.5; [−1.35 0.36]) |
| Hip circumference (cm) | 0.13 (1.25; [−0.40 2.89]) | 0.66 (−1.61; [−9.10 5.89]) | 0.74 (−0.14; [−0.95 0.68]) |
| WHR | 0.23 (0.01; [−0.01 0.03]) | 0.24 (0.04; [−0.03 0.11]) | 0.54 (0; [−0.01 0.01]) |
| FPG (mmol/L) | 0.32 (−0.35; [−1.04 0.35]) | 0.74 (−0.49; [−3.51 2.52]) | 0.55 (−0.08; [−0.33 0.18]) |
| FPI (pmol/L) | 0.93 (−1.11; [−26.53 24.31]) | 0.21 (−66.93; [−174.97 41.12]) | 0.3 (−3.57; [−10.40 3.25]) |
| HbA1c (%) | 0.3 (−0.23; [−0.67 0.21]) | 0.48 (−0.6; [−2.34 1.14]) | 0.7 (−0.03; [−0.19 0.13]) |
| Total Cholesterol (mmol/L) | 0.27 (−0.16; [−0.43 0.12]) | 0.62 (−0.27; [−1.37 0.84]) | 0.4 (−0.04; [−0.14 0.06]) |
| HDL-C (mmol/L) | 0.22 (−0.06; [−0.15 0.04]) | 0.93 (−0.02; [−0.39 0.35]) | 0.75 (0.01; [−0.03 0.04]) |
| LDL-C (mmol/L) | 0.38 (−0.1; [−0.34 0.14]) | 0.68 (−0.19; [−1.15 0.77]) | 0.25 (−0.05; [−0.14 0.04]) |
| Triglycerides (mmol/L) | 0.88 (−0.02; [−0.22 0.19]) | 0.13 (−0.58; [−1.36 0.20]) | 0.8 (−0.01; [−0.07 0.05]) |
| CrP (mg/L) | 0.74 (−0.26; [−1.77 1.26]) | 0.51 (2.23; [−4.65 9.11]) | 0.97 (0.01; [−0.70 0.73]) |
| Bilirubin (µg/mL) | 0.25 (0.79; [−0.56 2.14]) | 0.68 (1.24; [−4.98 7.47]) | 0.38 (−0.26; [−0.84 0.32]) |
| gammaGT (µkat/L) | 0.05 (0.16; [−0.002 0.33]) | 0.95 (−0.02; [−0.80 0.76]) | 0.2 (0.05; [−0.03 0.12]) |
| AP (µkat/L) | 0.45 (0.03; [−0.05 0.11]) | 0.73 (0.06; [−0.29 0.41]) | 0.07 (0.03; [−0.003 0.06]) |
| ASAT (µkat/L) | 0.33 (0.04; [−0.04 0.11]) | 0.44 (0.13; [−0.21 0.48]) | 0.83 (0; [−0.03 0.04]) |
| ALAT (µkat/L) | 0.7 (0.03; [−0.11 0.16]) | 0.93 (0.03; [−0.55 0.61]) | 0.54 (0.02; [−0.04 0.07]) |
| Steatosis (%) | 0.23 (0.02; [−0.02 0.06]) | 0.15 (0.13; [−0.05 0.31]) | 0.9 (0; [−0.01 0.01]) |
| Liver Fat Content (mean) | 0.19 (1.54; [−0.80 3.88]) | 0.003 (14.53; [5.06 23.99]) | 0.86 (−0.16; [−1.97 1.65]) |
| Presence of: | p-value adj (OR; [95% CI]) | p-value adj (OR; [95% CI]) | p-value adj (OR; [95% CI]) |
| NAFLD | N/A | N/A | N/A |
| NASH | 0.22 (1.34; [0.84 2.16]) | 0.01 (31.28; [1.87 522.09]) | 0.32 (0.86; [0.64 1.16]) |
| IGT | 0.03 (2.17; [1.10 4.31]) | 0.16 (5.97; [0.48 74.85]) | 0.35 (0.86; [0.63 1.18]) |
| Diabetes | 0.01 (2.16; [1.19 3.91]) | 0.07 (8.08; [0.84 77.46]) | 0.4 (0.88; [0.66 1.18]) |
| Hypertension | 0.06 (3.35; [0.97 11.61]) | 0.08 (312.17; [0.46 212,038.25]) | 0.99 (1; [0.80 1.25]) |
| Dyslipidemia | 0.5 (1.17; [0.74 1.84]) | 0.16 (4.49; [0.55 36.83]) | 0.93 (1.01; [0.79 1.30]) |
| rs4646404 G > A | p | padj | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | add | dom | rec | add | dom | rec | ||
| Total | lnPEMT sc | −0.6 (1.3) | −0.8 (1.4) | −0.9 (1.3) | 0.07 | 0.33 | 0.07 | 0.15 | 0.65 | 0.1 |
| lnPEMT vis | −1.3 (1.6) | −1.7 (1.6) | −1.4 (1.9) | 0.13 | 0.83 | 0.03 | 0.16 | 0.9 | 0.06 | |
| male | lnPEMT sc | −0.5 (1.3) | −0.8 (1.5) | −0.7 (0.9) | 0.35 | 0.9 | 0.26 | 0.24 | 0.62 | 0.2 |
| lnPEMT vis | −1.2 (1.6) | −1.4 (1.3) | −1.3 (1.5) | 0.73 | 0.96 | 0.69 | 0.19 | 0.85 | 0.49 | |
| female | lnPEMT sc | −0.6 (1.2) | −0.8 (1.3) | −1.0 (1.3) | 0.12 | 0.3 | 0.15 | 0.34 | 0.85 | 0.27 |
| lnPEMT vis | −1.3 (1.6) | −1.8 (1.6) | −1.4 (1.9) | 0.13 | 0.76 | 0.03 | 0.55 | 0.9 | 0.06 | |
| BMI < 30 kg/m2 | BMI 30–40 kg/m2 | BMI > 40 kg/m2 | |
|---|---|---|---|
| (N = 242) | (N = 383) | (N = 2301) | |
| Age (years) | 61 ± 15 | 49 ± 12 | 46 ± 11 |
| Female/Male (N) | 127/115 | 273/110 | 1632/669 |
| IGT/T2D (N) | 39 | 139 | 1026 |
| Body weight (kg) | 73 ± 11.2 | 104.4 ± 14.7 | 145.9 ± 26.1 |
| Height (m) | 1.7 ± 0.1 | 1.7 ± 0.1 | 1.7 ± 0.1 |
| BMI (kg/m2) | 25.3 ± 2.5 | 36.3 ± 2.8 | 50.3 ± 7.2 |
| Waist circumference (cm) | 90.9 ± 17.4 | 116.6 ± 14.8 | 142.4 ± 17.4 |
| Hip circumference (cm) | 95.3 ± 11.8 | 118.3 ± 12.9 | 148.6 ± 16.7 |
| WHR | 0.9 ± 0.1 | 1 ± 0.1 | 1 ± 0.1 |
| Body fat (%) | 23.9 ± 5.1 | 38 ± 8.8 | 48 ± 8.4 |
| FPG (mmol/L) | 5.7 ± 1.2 | 6.2 ± 2.6 | 6.6 ± 2.7 |
| FPI (pmol/L) | 56.7 ± 75.5 | 109.3 ± 143 | 151.8 ± 136.2 |
| HbA1c (%) | 5.6 ± 0.7 | 5.9 ± 0.9 | 6.2 ± 1.3 |
| Total cholesterol (mmol/L) | 5.2 ± 1 | 5.1 ± 1.2 | 4.9 ± 1.1 |
| HDL-C (mmol/L) | 1.4 ± 0.4 | 1.3 ± 0.4 | 1.2 ± 0.5 |
| LDL-C (mmol/L) | 3.1 ± 0.9 | 3.3 ± 1 | 3.1 ± 0.9 |
| Triglycerides (mmol/L) | 1.2 ± 0.6 | 1.7 ± 1.1 | 1.9 ± 1.4 |
| ALAT (µkat/L) | 0.5 ± 0.6 | 0.8 ± 2.8 | 0.8 ± 2.5 |
| ASAT (µkat/L) | 0.4 ± 0.2 | 0.6 ± 0.3 | 0.6 ± 0.3 |
| gamma-GT (µkat/L) | 1.2 ± 1.9 | 0.9 ± 1.8 | 1 ± 6.8 |
| Leptin (ng/mL) | 10.9 ± 10.7 | 31.6 ± 17.8 | 59.1 ± 34.4 |
| Sc/vis AT PEMT mRNA expression n (%) | 21 (4) | 48 (8) | 505 (88) |
| rs4646404 (G > A) carrier n (%) | 121 (10) | 137 (11) | 1011 (79) |
| NAFLD | NASH | p-Value | |
|---|---|---|---|
| (N = 29) | (N = 35) | ||
| Age (years) | 45.45 ± 10.13 | 47.26 ± 10.06 | 0.239 |
| Female/Male (N) | 24/5 | 21/14 | |
| IGT/T2D (N) | 4/8 | 4/22 | |
| Body weight (kg) | 130.28 ± 18.76 | 136.05 ± 18.82 | 0.222 |
| Height (m) | 1.7 ± 0.11 | 1.7 ± 0.09 | 0.914 |
| BMI (kg/m2) | 44.93 ± 5.06 | 46.97 ± 4.65 | 0.095 |
| Waist circumference (cm) | 125.08 ± 13.26 | 136.90 ± 12.37 | <0.001 |
| Hip circumference (cm) | 142.02 ± 11.8 | 140.25 ± 11.55 | 0.536 |
| WHR | 0.88 ± 0.08 | 0.98 ± 0.09 | <0.001 |
| Vis AT Volume (cm3) | 5152.00 ± 2273.73 | 7211.31 ± 2951.50 | 0.001 |
| Sc AT Volume (cm3) | 19,265.75 ± 3565.77 | 18,600.33 ± 2605.83 | 0.707 |
| Liver Volume (cm3) | 1949.47 ± 372.70 | 2544.66 ± 540.29 | <0.001 |
| FPG (mmol/L) | 5.19 ± 0.85 | 6.47 ± 2.29 | 0.010 |
| FPI (pmol/L) | 102.79 ± 74.70 | 119.13 ± 68.10 | 0.438 |
| HbA1c (%) | 5.21 ± 0.40 | 6.35 ± 1.51 | 0.001 |
| Total cholesterol (mmol/L) | 4.86 ± 0.97 | 4.35 ± 0.89 | 0.077 |
| HDL-C (mmol/L) | 1.25 ± 0.33 | 1.15 ± 0.25 | 0.268 |
| LDL-C (mmol/L) | 2.93 ± 0.86 | 2.51 ± 0.82 | 0.095 |
| Triglycerides (mmol/L) | 1.4 ± 0.64 | 1.48 ± 0.80 | 0.750 |
| gammaGT (µkat/L) | 0.65 ± 0.84 | 0.75 ± 0.55 | 0.594 |
| AP (µkat/L) | 1.32 ± 0.37 | 1.20 ± 0.30 | 0.174 |
| ASAT (µkat/L) | 0.59 ± 0.28 | 0.76 ± 0.34 | 0.041 |
| ALAT (µkat/L) | 0.78 ± 0.48 | 1.07 ± 0.58 | 0.046 |
| Bilirubin (µg/mL) | 9.08 ± 6.22 | 9.85 ± 5.48 | 0.610 |
| CRP (mg/L) | 8.26 ± 7.05 | 8.40 ± 7.87 | 0.943 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, C.; Holstein, D.J.F.; Garcia-Cubero, N.; Moulla, Y.; Stroh, C.; Dietrich, A.; Schön, M.R.; Gärtner, D.; Lohmann, T.; Dressler, M.; et al. The Role of Phosphatidylethanolamine N-Methyltransferase (PEMT) and Its Waist-Hip-Ratio-Associated Locus rs4646404 in Obesity-Related Metabolic Traits and Liver Disease. Int. J. Mol. Sci. 2023, 24, 16850. https://doi.org/10.3390/ijms242316850
Sun C, Holstein DJF, Garcia-Cubero N, Moulla Y, Stroh C, Dietrich A, Schön MR, Gärtner D, Lohmann T, Dressler M, et al. The Role of Phosphatidylethanolamine N-Methyltransferase (PEMT) and Its Waist-Hip-Ratio-Associated Locus rs4646404 in Obesity-Related Metabolic Traits and Liver Disease. International Journal of Molecular Sciences. 2023; 24(23):16850. https://doi.org/10.3390/ijms242316850
Chicago/Turabian StyleSun, Chang, David J. F. Holstein, Natalia Garcia-Cubero, Yusef Moulla, Christine Stroh, Arne Dietrich, Michael R. Schön, Daniel Gärtner, Tobias Lohmann, Miriam Dressler, and et al. 2023. "The Role of Phosphatidylethanolamine N-Methyltransferase (PEMT) and Its Waist-Hip-Ratio-Associated Locus rs4646404 in Obesity-Related Metabolic Traits and Liver Disease" International Journal of Molecular Sciences 24, no. 23: 16850. https://doi.org/10.3390/ijms242316850