Coordinated Transcriptional Waves Define the Inflammatory Response of Primary Microglial Culture
Abstract
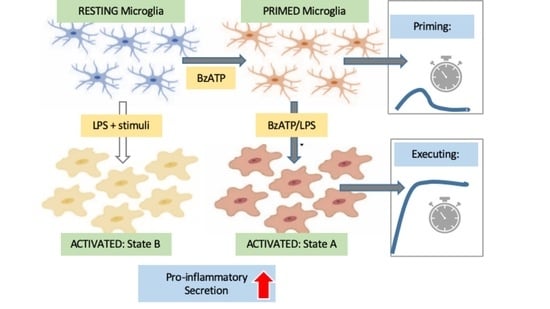
:1. Introduction
2. Results
2.1. Functional Response of Primary Microglia
2.2. A Short-Lived Transient Wave of Gene Expression by bzATP
2.3. Global Alteration of the Cell Transcriptome by bzATP/LPS Is Synchronized and Long-Lasting
2.4. Expression of Interleukins as Indicators of Microglial Response
2.5. Changes in Gene Expression by Orders of Magnitude Drive the Cellular State
2.6. Enrichment Analysis of the Transcriptome Induced by bzATP/LPS
2.7. Inflammatory-Related Signaling Pathways Are Significantly Induced by Exposing Cells to LPS
2.8. Alternative Signaling Patterns Dominate the Microglia Activation States
2.9. Stimulation of Microglia by bzATP/LPS Affects the Expression of Abundant ncRNAs
3. Discussion
4. Materials and Methods
4.1. Compounds and Reagents
4.2. Preparation of Microglial Cultures
4.3. Measurement of Cytokines
4.4. RNA-Seq
4.5. Gel Based RT-PCR
4.6. Bioinformatic Analysis and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| ATP | Adenosine tri-phosphate |
| BSA | Bovine serum albumin |
| CNS | Central nervous system |
| DE | Differentially expressed |
| DMEM | Dulbecco’s modified Eagle medium |
| FC | Fold change |
| FDR | False discovery rate |
| GO | Gene ontology |
| h | Hours |
| IL | Interleukin |
| IFNγ | Interferon gamma |
| LPS | Lipopolysaccharide |
| NT | Not treated |
| PCA | Principal component analysis |
| RNA-seq | RNA sequencing analysis |
| TMM | Trimmed mean of means |
References
- Amor, S.; Peferoen, L.A.; Vogel, D.Y.; Breur, M.; van der Valk, P.; Baker, D.; van Noort, J.M. Inflammation in neurodegenerative diseases—An update. Immunology 2014, 142, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Clayton, K.A.; Van Enoo, A.A.; Ikezu, T. Alzheimer’s disease: The role of microglia in brain homeostasis and proteopathy. Front. Neurosci. 2017, 11, 680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.-M.; Hong, J.-S. Why neurodegenerative diseases are progressive: Uncontrolled inflammation drives disease progression. Trends Immunol. 2008, 29, 357–365. [Google Scholar] [CrossRef] [Green Version]
- Wolf, S.A.; Boddeke, H.W.; Kettenmann, H. Microglia in Physiology and Disease. Annu. Rev. Physiol. 2017, 79, 619–643. [Google Scholar] [CrossRef] [PubMed]
- Imura, Y.; Morizawa, Y.; Komatsu, R.; Shibata, K.; Shinozaki, Y.; Kasai, H.; Moriishi, K.; Moriyama, Y.; Koizumi, S. Microglia release ATP by exocytosis. Glia 2013, 61, 1320–1330. [Google Scholar] [CrossRef]
- Sanz, J.M.; Di Virgilio, F. Kinetics and mechanism of ATP-dependent IL-1β release from microglial cells. J. Immunol. 2000, 164, 4893–4898. [Google Scholar] [CrossRef] [Green Version]
- Neher, J.J.; Neniskyte, U.; Brown, G.C. Primary phagocytosis of neurons by inflamed microglia: Potential roles in neurodegeneration. Front. Pharm. 2012, 3, 27. [Google Scholar] [CrossRef] [Green Version]
- Fricker, M.; Vilalta, A.; Tolkovsky, A.M.; Brown, G.C. Caspase inhibitors protect neurons by enabling selective necroptosis of inflamed microglia. J. Biol. Chem. 2013, 288, 9145–9152. [Google Scholar] [CrossRef] [Green Version]
- Hammond, T.R.; Dufort, C.; Dissing-Olesen, L.; Giera, S.; Young, A.; Wysoker, A.; Walker, A.J.; Gergits, F.; Segel, M.; Nemesh, J. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 2019, 50, 253–271.e6. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.Y.; Tan, M.S.; Yu, J.T.; Tan, L. Role of pro-inflammatory cytokines released from microglia in Alzheimer’s disease. Ann. Transl. Med. 2015, 3, 136. [Google Scholar] [CrossRef]
- Rodriguez-Gomez, J.A.; Kavanagh, E.; Engskog-Vlachos, P.; Engskog, M.K.R.; Herrera, A.J.; Espinosa-Oliva, A.M.; Joseph, B.; Hajji, N.; Venero, J.L.; Burguillos, M.A. Microglia: Agents of the CNS Pro-Inflammatory Response. Cells 2020, 9, 1717. [Google Scholar] [CrossRef] [PubMed]
- Shoham, S.; Linial, M.; Weinstock, M. Age-Induced Spatial Memory Deficits in Rats Are Correlated with Specific Brain Region Alterations in Microglial Morphology and Gene Expression. J. Neuroimmune Pharm. 2019, 14, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Shemer, A.; Erny, D.; Jung, S.; Prinz, M. Microglia plasticity during health and disease: An immunological perspective. Trends Immunol. 2015, 36, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Dissing-Olesen, L.; MacVicar, B.A.; Stevens, B. Microglia: Dynamic mediators of synapse development and plasticity. Trends Immunol. 2015, 36, 605–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickman, S.; Izzy, S.; Sen, P.; Morsett, L.; El Khoury, J. Microglia in neurodegeneration. Nat. Neurosci. 2018, 21, 1359–1369. [Google Scholar] [CrossRef]
- Linial, M.; Stern, A.; Weinstock, M. Effect of ladostigil treatment of aging rats on gene expression in four brain areas associated with regulation of memory. Neuropharmacology 2020, 177, 108229. [Google Scholar] [CrossRef]
- Hide, I.; Tanaka, M.; Inoue, A.; Nakajima, K.; Kohsaka, S.; Inoue, K.; Nakata, Y. Extracellular ATP triggers tumor necrosis factor-alpha release from rat microglia. J. Neurochem. 2000, 75, 965–972. [Google Scholar] [CrossRef]
- Illes, P. P2X7 Receptors Amplify CNS Damage in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 5996. [Google Scholar] [CrossRef]
- Lambertsen, K.L.; Clausen, B.H.; Babcock, A.A.; Gregersen, R.; Fenger, C.; Nielsen, H.H.; Haugaard, L.S.; Wirenfeldt, M.; Nielsen, M.; Dagnaes-Hansen, F. Microglia protect neurons against ischemia by synthesis of tumor necrosis factor. J. Neurosci. 2009, 29, 1319–1330. [Google Scholar] [CrossRef] [Green Version]
- Cianciulli, A.; Porro, C.; Calvello, R.; Trotta, T.; Lofrumento, D.D.; Panaro, M.A. Microglia Mediated Neuroinflammation: Focus on PI3K Modulation. Biomolecules 2020, 10, 137. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Min, J.-S.; Kim, B.; Chae, U.-B.; Yun, J.W.; Choi, M.-S.; Kong, I.-K.; Chang, K.-T.; Lee, D.-S. Mitochondrial ROS govern the LPS-induced pro-inflammatory response in microglia cells by regulating MAPK and NF-κB pathways. Neurosci. Lett. 2015, 584, 191–196. [Google Scholar] [CrossRef]
- Zhou, L.-t.; Wang, K.-j.; Li, L.; Li, H.; Geng, M. Pinocembrin inhibits lipopolysaccharide-induced inflammatory mediators production in BV2 microglial cells through suppression of PI3K/Akt/NF-κB pathway. Eur. J. Pharmacol. 2015, 761, 211–216. [Google Scholar] [CrossRef]
- Akhmetzyanova, E.; Kletenkov, K.; Mukhamedshina, Y.; Rizvanov, A. Different Approaches to Modulation of Microglia Phenotypes After Spinal Cord Injury. Front. Syst. Neurosci. 2019, 13, 37. [Google Scholar] [CrossRef] [Green Version]
- Hirbec, H.E.; Noristani, H.N.; Perrin, F.E. Microglia responses in acute and chronic neurological diseases: What microglia-specific transcriptomic studies taught (and did not teach) us. Front. Aging Neurosci. 2017, 9, 227. [Google Scholar] [CrossRef] [Green Version]
- Timmerman, R.; Burm, S.M.; Bajramovic, J.J. An Overview of in vitro Methods to Study Microglia. Front. Cell Neurosci. 2018, 12, 242. [Google Scholar] [CrossRef]
- Lively, S.; Schlichter, L.C. Microglia Responses to Pro-inflammatory Stimuli (LPS, IFNγ+TNFα) and Reprogramming by Resolving Cytokines (IL-4, IL-10). Front. Cell Neurosci. 2018, 12, 215. [Google Scholar] [CrossRef] [Green Version]
- Douvaras, P.; Sun, B.; Wang, M.; Kruglikov, I.; Lallos, G.; Zimmer, M.; Terrenoire, C.; Zhang, B.; Gandy, S.; Schadt, E.; et al. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Rep. 2017, 8, 1516–1524. [Google Scholar] [CrossRef] [Green Version]
- McQuade, A.; Coburn, M.; Tu, C.H.; Hasselmann, J.; Davtyan, H.; Blurton-Jones, M. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol. Neurodegener. 2018, 13, 67. [Google Scholar] [CrossRef] [Green Version]
- Welser-Alves, J.V.; Milner, R. Microglia are the major source of TNF-α and TGF-β1 in postnatal glial cultures; regulation by cytokines, lipopolysaccharide, and vitronectin. Neurochem. Int. 2013, 63, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Bohlen, C.J.; Bennett, F.C.; Tucker, A.F.; Collins, H.Y.; Mulinyawe, S.B.; Barres, B.A. Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures. Neuron 2017, 94, 759–773.e8. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; He, H.; Mo, L.; Liu, Q.; Yang, F.; Zhou, Y.; Li, L.; Su, D.; Yi, S.; Zhang, J. Mapping the Plasticity of Morphology, Molecular Properties and Function in Mouse Primary Microglia. Front. Cell Neurosci. 2021, 15, 811061. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.S.A.; Oliver, P.L. ROS Generation in Microglia: Understanding Oxidative Stress and Inflammation in Neurodegenerative Disease. Antioxidants 2020, 9, 743. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A.; Das, A.; Ray, S.K.; Banik, N.L. Role of pro-inflammatory cytokines released from microglia in neurodegenerative diseases. Brain Res. Bull. 2012, 87, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.C.; Hu, S.; Close, K.; Choi, C.S.; Molitor, T.W.; Novick, W.J.; Peterson, P.K. Cytokine release from microglia: Differential inhibition by pentoxifylline and dexamethasone. J. Infect. Dis. 1992, 166, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014, 43, D447–D452. [Google Scholar] [CrossRef] [Green Version]
- Martens, M.; Ammar, A.; Riutta, A.; Waagmeester, A.; Slenter, D.N.; Hanspers, K.; Miller, R.A.; Digles, D.; Lopes, E.N.; Ehrhart, F.; et al. WikiPathways: Connecting communities. Nucleic Acids Res. 2021, 49, D613–D621. [Google Scholar] [CrossRef]
- Zhang, J.D.; Wiemann, S. KEGGgraph: A graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics 2009, 25, 1470–1471. [Google Scholar] [CrossRef] [Green Version]
- Mosher, K.I.; Wyss-Coray, T. Microglial dysfunction in brain aging and Alzheimer’s disease. Biochem. Pharmacol. 2014, 88, 594–604. [Google Scholar] [CrossRef] [Green Version]
- McInturff, J.E.; Modlin, R.L.; Kim, J. The role of toll-like receptors in the pathogenesis and treatment of dermatological disease. J. Investig. Dermatol. 2005, 125, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.A. The multifaceted profile of activated microglia. Mol. Neurobiol. 2009, 40, 139–156. [Google Scholar] [CrossRef]
- Suuronen, T.; Nuutinen, T.; Huuskonen, J.; Ojala, J.; Thornell, A.; Salminen, A. Anti-inflammatory effect of selective estrogen receptor modulators (SERMs) in microglial cells. Inflamm. Res. 2005, 54, 194–203. [Google Scholar] [CrossRef]
- York, E.M.; Bernier, L.P.; MacVicar, B.A. Microglial modulation of neuronal activity in the healthy brain. Dev. Neurobiol. 2018, 78, 593–603. [Google Scholar] [CrossRef]
- Raouf, R.; Chabot-Doré, A.-J.; Ase, A.R.; Blais, D.; Séguéla, P. Differential regulation of microglial P2X4 and P2X7 ATP receptors following LPS-induced activation. Neuropharmacology 2007, 53, 496–504. [Google Scholar] [CrossRef]
- Gulke, E.; Gelderblom, M.; Magnus, T. Danger signals in stroke and their role on microglia activation after ischemia. Adv. Neurol. Disord. 2018, 11, 1756286418774254. [Google Scholar] [CrossRef] [Green Version]
- Kiyota, T.; Gendelman, H.E.; Weir, R.A.; Higgins, E.E.; Zhang, G.; Jain, M. CCL2 affects β-amyloidosis and progressive neurocognitive dysfunction in a mouse model of Alzheimer’s disease. Neurobiol. Aging 2013, 34, 1060–1068. [Google Scholar] [CrossRef] [Green Version]
- Joly-Amado, A.; Hunter, J.; Quadri, Z.; Zamudio, F.; Rocha-Rangel, P.V.; Chan, D.; Kesarwani, A.; Nash, K.; Lee, D.C.; Morgan, D. CCL2 overexpression in the brain promotes glial activation and accelerates tau pathology in a mouse model of tauopathy. Front. Immunol. 2020, 11, 997. [Google Scholar] [CrossRef]
- Elling, R.; Robinson, E.K.; Shapleigh, B.; Liapis, S.C.; Covarrubias, S.; Katzman, S.; Groff, A.F.; Jiang, Z.; Agarwal, S.; Motwani, M.; et al. Genetic Models Reveal cis and trans Immune-Regulatory Activities for lincRNA-Cox2. Cell Rep. 2018, 25, 1511–1524.e6. [Google Scholar] [CrossRef] [Green Version]
- Heyen, J.R.; Ye, S.-m.; Finck, B.N.; Johnson, R.W. Interleukin (IL)-10 inhibits IL-6 production in microglia by preventing activation of NF-κB. Mol. Brain Res. 2000, 77, 138–147. [Google Scholar] [CrossRef]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef] [Green Version]
- Henn, A.; Lund, S.; Hedtjärn, M.; Schrattenholz, A.; Pörzgen, P.; Leist, M. The suitability of BV2 cells as alternative model system for primary microglia cultures or for animal experiments examining brain inflammation. ALTEX Altern. Anim. Exp. 2009, 26, 83–94. [Google Scholar] [CrossRef] [Green Version]
- Mendonca, P.; Taka, E.; Bauer, D.; Cobourne-Duval, M.; Soliman, K.F. The attenuating effects of 1, 2, 3, 4, 6 penta-O-galloyl-β-d-glucose on inflammatory cytokines release from activated BV-2 microglial cells. J. Neuroimmunol. 2017, 305, 9–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.; Kim, S.H.; Arifuzzaman, S.; Yoon, T.; Chai, J.C.; Lee, Y.S.; Park, K.S.; Jung, K.H.; Chai, Y.G. Transcriptome sequencing reveals that LPS-triggered transcriptional responses in established microglia BV2 cell lines are poorly representative of primary microglia. J. Neuroinflamm. 2016, 13, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirbec, H.; Marmai, C.; Duroux-Richard, I.; Roubert, C.; Esclangon, A.; Croze, S.; Lachuer, J.; Peyroutou, R.; Rassendren, F. The microglial reaction signature revealed by RNAseq from individual mice. Glia 2018, 66, 971–986. [Google Scholar] [CrossRef] [PubMed]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. [Google Scholar] [CrossRef] [Green Version]
- Pulido-Salgado, M.; Vidal-Taboada, J.M.; Barriga, G.G.; Sola, C.; Saura, J. RNA-Seq transcriptomic profiling of primary murine microglia treated with LPS or LPS + IFNγ. Sci. Rep. 2018, 8, 16096. [Google Scholar] [CrossRef] [Green Version]
- Gerrits, E.; Heng, Y.; Boddeke, E.; Eggen, B.J.L. Transcriptional profiling of microglia; current state of the art and future perspectives. Glia 2020, 68, 740–755. [Google Scholar] [CrossRef] [Green Version]
- Chhor, V.; Le Charpentier, T.; Lebon, S.; Ore, M.V.; Celador, I.L.; Josserand, J.; Degos, V.; Jacotot, E.; Hagberg, H.; Savman, K.; et al. Characterization of phenotype markers and neuronotoxic potential of polarised primary microglia in vitro. Brain Behav. Immun. 2013, 32, 70–85. [Google Scholar] [CrossRef]
- Wong, E.T.; Tergaonkar, V. Roles of NF-κB in health and disease: Mechanisms and therapeutic potential. Clin. Sci. 2009, 116, 451–465. [Google Scholar] [CrossRef]
- Weinstock, M.; Bejar, C.; Schorer-Apelbaum, D.; Panarsky, R.; Luques, L.; Shoham, S. Dose-dependent effects of ladostigil on microglial activation and cognition in aged rats. J. Neuroimmune Pharmacol. 2013, 8, 345–355. [Google Scholar] [CrossRef]
- Panarsky, R.; Luques, L.; Weinstock, M. Anti-inflammatory effects of ladostigil and its metabolites in aged rat brain and in microglial cells. J. Neuroimmune Pharmacol. 2012, 7, 488–498. [Google Scholar] [CrossRef]
- Reichert, F.; Rotshenker, S. Complement-receptor-3 and scavenger-receptor-AI/II mediated myelin phagocytosis in microglia and macrophages. Neurobiol. Dis. 2003, 12, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Gitik, M.; Liraz-Zaltsman, S.; Oldenborg, P.A.; Reichert, F.; Rotshenker, S. Myelin down-regulates myelin phagocytosis by microglia and macrophages through interactions between CD47 on myelin and SIRPα (signal regulatory protein-alpha) on phagocytes. J. Neuroinflamm. 2011, 8, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shamash, S.; Reichert, F.; Rotshenker, S. The cytokine network of Wallerian degeneration: Tumor necrosis factor-alpha, interleukin-1α, and interleukin-1β. J. Neurosci. 2002, 22, 3052–3060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zohar, K.; Lezmi, E.; Eliyahu, T.; Linial, M. Ladostigil Attenuates Induced Oxidative Stress in Human Neuroblast-like SH-SY5Y Cells. Biomedicines 2021, 9, 1251. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef] [Green Version]







| FC1 3 h vs. N.T. | Trend FC1 a | FC2 8 h vs. 3 h | Trend FC2 | Combined Trend |
|---|---|---|---|---|
| log(FC1) > 0.5 | Up | log(FC2) ≥ log(FC1) + 0.5 | Up | Up–Up |
| log(FC1) > 0.5 | Up | log(FC2) ≤ log(FC1) + 0.5 and log(FC2) ≥ log(FC1) − 0.5 | Same | Up–Same |
| log(FC1) > 0.5 | Up | log(FC2) < log(FC1) − 0.5 | Down | Up–Down |
| −0.5 ≤ log(FC1) ≤ 0.5 | Same | log(FC2) > log(FC1) + 0.5 | Up | Same–Up |
| −0.5 ≤ log(FC1) ≤ 0.5 | Same | log(FC2) ≤ log(FC1) + 0.5 and log(FC2) ≥ log(FC1) − 0.5 | Same | Same–Same |
| −0.5 ≤ log(FC1) ≤ 0.5 | Same | log(FC2) < log(FC1) − 0.5 | Down | Same–Down |
| log(FC1) < −0.5 | Down | log(FC2) ≥ log(FC1) + 0.5 | Up | Down–Up |
| log(FC1) < −0.5 | Down | log(FC2) ≤ log(FC1) + 0.5 and log(FC2) ≥ log(FC1) − 0.5 | Same | Down–Same |
| log(FC1) < −0.5 | Down | log(FC2) < log(FC1) − 0.5 | Down | Down–Down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zohar, K.; Lezmi, E.; Reichert, F.; Eliyahu, T.; Rotshenker, S.; Weinstock, M.; Linial, M. Coordinated Transcriptional Waves Define the Inflammatory Response of Primary Microglial Culture. Int. J. Mol. Sci. 2023, 24, 10928. https://doi.org/10.3390/ijms241310928
Zohar K, Lezmi E, Reichert F, Eliyahu T, Rotshenker S, Weinstock M, Linial M. Coordinated Transcriptional Waves Define the Inflammatory Response of Primary Microglial Culture. International Journal of Molecular Sciences. 2023; 24(13):10928. https://doi.org/10.3390/ijms241310928
Chicago/Turabian StyleZohar, Keren, Elyad Lezmi, Fanny Reichert, Tsiona Eliyahu, Shlomo Rotshenker, Marta Weinstock, and Michal Linial. 2023. "Coordinated Transcriptional Waves Define the Inflammatory Response of Primary Microglial Culture" International Journal of Molecular Sciences 24, no. 13: 10928. https://doi.org/10.3390/ijms241310928