Investigating the Physiological and Molecular Responses of Solanum lycopersicum hp Mutants to Light of Different Quality for Biotechnological Applications
Abstract
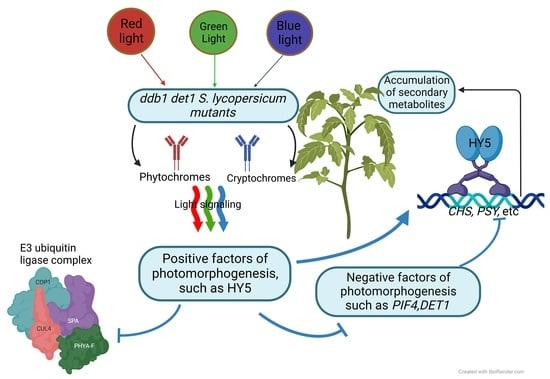
:1. Introduction
2. Results
2.1. Low-Molecular Weight Antioxidant Capacity, Total Phenolic, and Flavonoid Contents
2.2. CO2 Gas Exchange, Transpiration, and Dry Weight Percentage
2.3. Photochemical Activity
2.4. Gene Expression
2.5. Scanning Electron Microscopy
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Experimental Design
4.2. Low-Molecular-Weight Antioxidants
4.3. CO2 Gas Exchange and Transpiration
4.4. Photochemical Activity
4.5. RNA Extraction and RT–PCR
4.6. Scanning Electron Microscopy
4.7. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Samtiya, M.; Aluko, R.E.; Dhewa, T.; Moreno-Rojas, J.M. Potential Health Benefits of Plant Food-Derived Bioactive Components: An Overview. Foods 2021, 10, 839. [Google Scholar] [CrossRef]
- Ouzounis, T.; Rosenqvist, E.; Ottosen, C.-O. Spectral Effects of Artificial Light on Plant Physiology and Secondary Metabolism: A Review. HortScience 2015, 50, 1128–1135. [Google Scholar] [CrossRef]
- Pashkovskiy, P.; Kreslavski, V.D.; Ivanov, Y.; Ivanova, A.; Kartashov, A.; Shmarev, A.; Strokina, V.; Kuznetsov, V.V.; Allakhverdiev, S.I. Influence of Light of Different Spectral Compositions on the Growth, Photosynthesis, and Expression of Light-Dependent Genes of Scots Pine Seedlings. Cells 2021, 10, 3284. [Google Scholar] [CrossRef]
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The Effect of Developmental and Environmental Factors on Secondary Metabolites in Medicinal Plants. Plant Physiol. Biochem. 2020, 148, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, P.; Yang, T.-J.; Song, Y.H. Effect of Far-Red Light on the Production and Diversity of Ginsenosides in Leaves of Panax Ginseng Meyer. Appl. Biol. Chem. 2023, 66, 16. [Google Scholar] [CrossRef]
- Pashkovskiy, P.; Kreslavski, V.; Khudyakova, A.; Ashikhmin, A.; Bolshakov, M.; Kozhevnikova, A.; Kosobryukhov, A.; Kuznetsov, V.V.; Allakhverdiev, S.I. Effect of High-Intensity Light on the Photosynthetic Activity, Pigment Content and Expression of Light-Dependent Genes of Photomorphogenetic Solanum lycopersicum Hp Mutants. Plant Physiol. Biochem. 2021, 167, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, L.; Zou, H.; Qiu, L.; Zheng, Y.; Yang, D.; Wang, Y. Effects of Light on Secondary Metabolite Biosynthesis in Medicinal Plants. Front. Plant Sci. 2021, 12, 781236. [Google Scholar] [CrossRef]
- Khansari, N.; Shakiba, Y.; Mahmoudi, M. Chronic Inflammation and Oxidative Stress as a Major Cause of Age-Related Diseases and Cancer. Recent Pat. Inflamm. Allergy Drug Discov. 2009, 3, 73–80. [Google Scholar] [CrossRef]
- Gutiérrez-Lomelí, M.; Del Toro-Sánchez, C.L.; Rodríguez-Sahagún, A.; Castellanos-Hernández, O.A. Natural Products Extracts: Terpenes and Phenolics. Biotechnol. Prod. Plant Second. Metab. 2012, 21, 21–35. [Google Scholar]
- Dixon, R.A.; Lamb, C.J.; Masoud, S.; Sewalt, V.J.; Paiva, N.L. Metabolic Engineering: Prospects for Crop Improvement through the Genetic Manipulation of Phenylpropanoid Biosynthesis and Defense Responses—A Review. Gene 1996, 179, 61–71. [Google Scholar] [CrossRef]
- Pashkovskiy, P.; Vereshchagin, M.; Kreslavski, V.; Ivanov, Y.; Kumachova, T.; Ryabchenko, A.; Voronkov, A.; Kosobryukhov, A.; Kuznetsov, V.; Allakhverdiev, S.I. Effect of Phytochrome Deficiency on Photosynthesis, Light-Related Genes Expression and Flavonoid Accumulation in Solanum lycopersicum under Red and Blue Light. Cells 2022, 11, 3437. [Google Scholar] [CrossRef]
- Thoma, F.; Somborn-Schulz, A.; Schlehuber, D.; Keuter, V.; Deerberg, G. Effects of Light on Secondary Metabolites in Selected Leafy Greens: A Review. Front. Plant Sci. 2020, 11, 497. [Google Scholar] [CrossRef] [PubMed]
- Kreslavski, V.D.; Los, D.A.; Schmitt, F.-J.; Zharmukhamedov, S.K.; Kuznetsov, V.V.; Allakhverdiev, S.I. The Impact of the Phytochromes on Photosynthetic Processes. Biochim. Biophys. Acta (BBA)-Bioenerg. 2018, 1859, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deng, H.; Miao, M.; Li, H.; Huang, S.; Wang, S.; Liu, Y. Tomato MBD 5, a Methyl CpG Binding Domain Protein, Physically Interacting with UV-Damaged DNA Binding Protein-1, Functions in Multiple Processes. New Phytol. 2016, 210, 208–226. [Google Scholar] [CrossRef] [PubMed]
- Caspi, N.; Levin, I.; Chamovitz, D.A.; Reuveni, M. A Mutation in the Tomato DDB1 Gene Affects Cell and Chloroplast Compartment Size and CDT1 Transcript. Plant Signal. Behav. 2008, 3, 641–649. [Google Scholar] [CrossRef]
- Levin, I.; De Vos, C.R.; Tadmor, Y.; Bovy, A.; Lieberman, M.; Oren-Shamir, M.; Segev, O.; Kolotilin, I.; Keller, M.; Ovadia, R. High Pigment Tomato Mutants—More than Just Lycopene (a Review). Isr. J. Plant Sci. 2006, 54, 179–190. [Google Scholar] [CrossRef]
- Liu, C.-C.; Chi, C.; Jin, L.-J.; Zhu, J.; Yu, J.-Q.; Zhou, Y.-H. The BZip Transcription Factor HY5 Mediates CRY1a-Induced Anthocyanin Biosynthesis in Tomato. Plant Cell Environ. 2018, 41, 1762–1775. [Google Scholar] [CrossRef]
- Shin, J.; Park, E.; Choi, G. PIF3 Regulates Anthocyanin Biosynthesis in an HY5-Dependent Manner with Both Factors Directly Binding Anthocyanin Biosynthetic Gene Promoters in Arabidopsis. Plant J. 2007, 49, 981–994. [Google Scholar] [CrossRef]
- Wang, A.; Chen, D.; Ma, Q.; Rose, J.K.; Fei, Z.; Liu, Y.; Giovannoni, J.J. The Tomato High Pigment1/Damaged DNA Binding Protein 1 Gene Contributes to Regulation of Fruit Ripening. Hortic. Res. 2019, 6, 15. [Google Scholar] [CrossRef]
- Pepper, A.; Delaney, T.; Washburnt, T.; Poole, D.; Chory, J. DET1, a Negative Regulator of Light-Mediated Development and Gene Expression in Arabidopsis, Encodes a Novel Nuclear-Localized Protein. Cell 1994, 78, 109–116. [Google Scholar] [CrossRef]
- Lieberman, M.; Segev, O.; Gilboa, N.; Lalazar, A.; Levin, I. The Tomato Homolog of the Gene Encoding UV-Damaged DNA Binding Protein 1 (DDB1) Underlined as the Gene That Causes the High Pigment-1 Mutant Phenotype. Theor. Appl. Genet. 2004, 108, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Kilambi, H.V.; Kumar, R.; Sharma, R.; Sreelakshmi, Y. Chromoplast-Specific Carotenoid-Associated Protein Appears to Be Important for Enhanced Accumulation of Carotenoids in Hp1 Tomato Fruits. Plant Physiol. 2013, 161, 2085–2101. [Google Scholar] [CrossRef]
- Azari, R.; Reuveni, M.; Evenor, D.; Nahon, S.; Shlomo, H.; Chen, L.; Levin, I. Overexpression of UV-Damaged DNA Binding Protein 1 Links Plant Development and Phytonutrient Accumulation in High Pigment-1 Tomato. J. Exp. Bot. 2010, 61, 3627–3637. [Google Scholar] [CrossRef] [PubMed]
- Fernando, V.C.; Schroeder, D. Shedding Light on Plant Development: Light Signalling in the Model Plant Arabidopsis thaliana. Ceylon J. Sci. 2016, 45, 3–13. [Google Scholar] [CrossRef]
- Kapoor, L.; Simkin, A.J.; George Priya Doss, C.; Siva, R. Fruit Ripening: Dynamics and Integrated Analysis of Carotenoids and Anthocyanins. BMC Plant Biol. 2022, 22, 27. [Google Scholar] [CrossRef] [PubMed]
- Yadukrishnan, P.; Datta, S. Light and Abscisic Acid Interplay in Early Seedling Development. New Phytol. 2021, 229, 763–769. [Google Scholar] [CrossRef]
- Nassrallah, A.; Rougée, M.; Bourbousse, C.; Drevensek, S.; Fonseca, S.; Iniesto, E.; Ait-Mohamed, O.; Deton-Cabanillas, A.-F.; Zabulon, G.; Ahmed, I. DET1-Mediated Degradation of a SAGA-like Deubiquitination Module Controls H2Bub Homeostasis. eLife 2018, 7, e37892. [Google Scholar] [CrossRef]
- Lau, O.S.; Deng, X.W. The Photomorphogenic Repressors COP1 and DET1: 20 Years Later. Trends Plant Sci. 2012, 17, 584–593. [Google Scholar] [CrossRef]
- Bernhardt, A.; Lechner, E.; Hano, P.; Schade, V.; Dieterle, M.; Anders, M.; Dubin, M.J.; Benvenuto, G.; Bowler, C.; Genschik, P. CUL4 Associates with DDB1 and DET1 and Its Downregulation Affects Diverse Aspects of Development in Arabidopsis thaliana. Plant J. 2006, 47, 591–603. [Google Scholar] [CrossRef]
- Dieterle, M.; Zhou, Y.-C.; Schäfer, E.; Funk, M.; Kretsch, T. EID1, an F-Box Protein Involved in Phytochrome A-Specific Light Signaling. Genes Dev. 2001, 15, 939–944. [Google Scholar] [CrossRef]
- Lytovchenko, A.; Eickmeier, I.; Pons, C.; Osorio, S.; Szecowka, M.; Lehmberg, K.; Arrivault, S.; Tohge, T.; Pineda, B.; Anton, M.T. Tomato Fruit Photosynthesis Is Seemingly Unimportant in Primary Metabolism and Ripening but Plays a Considerable Role in Seed Development. Plant Physiol. 2011, 157, 1650–1663. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant Activity Applying an Improved ABTS Radical Cation Decolorization Assay. Free. Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Singleton, V.L.; Rossi, J.A. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
- Kim, D.-O.; Chun, O.K.; Kim, Y.J.; Moon, H.-Y.; Lee, C.Y. Quantification of Polyphenolics and Their Antioxidant Capacity in Fresh Plums. J. Agric. Food Chem. 2003, 51, 6509–6515. [Google Scholar] [CrossRef] [PubMed]
- Stetsenko, L.A.; Pashkovsky, P.P.; Voloshin, R.A.; Kreslavski, V.D.; Kuznetsov, V.V.; Allakhverdiev, S.I. Role of Anthocyanin and Carotenoids in the Adaptation of the Photosynthetic Apparatus of Purple-and Green-Leaved Cultivars of Sweet Basil (Ocimum basilicum) to High-Intensity Light. Photosynthetica 2020, 58, 890–901. [Google Scholar] [CrossRef]



| Variant | Light | TEAC | Total Phenols | Flavonoids |
|---|---|---|---|---|
| WT | WFL | 9.17 ± 0.12 b | 0.83 ± 0.02 b | 0.98 ± 0.02 a |
| 3005 | 12.80 ± 1.03 a | 1.19 ± 0.1 a | 1.42 ± 0.12 a | |
| 4012 | 9.53 ± 0.38 b | 1.06 ± 0.05 ab | 1.03 ± 0.01 a | |
| 0279 | 12.17 ± 0.19 a | 0.98 ± 0.02 b | 1.14 ± 0.12 a | |
| 3538 | 11.07 ± 0.73 ab | 0.95 ± 0.02 b | 1.12 ± 0.13 a | |
| WT | WL | 7.27 ± 0.84 c | 0.58 ± 0.07 c | 0.67 ± 0.09 c |
| 3005 | 14.20 ± 0.76 a | 1.32 ± 0.02 a | 1.37 ± 0.03 a | |
| 4012 | 8.10 ± 0.32 bc | 0.74 ± 0.04 bc | 0.72 ± 0.06 c | |
| 0279 | 6.03 ± 0.47 c | 0.39 ± 0.02 d | 0.4 ± 0.04 d | |
| 3538 | 9.90 ± 0.42 b | 0.91 ± 0.09 b | 1.02 ± 0.04 b | |
| WT | RL | 9.80 ± 0.46 c | 0.98 ± 0.03 c | 0.98 ± 0.08 c |
| 3005 | 19.97 ± 0.26 a | 2.16 ± 0.15 a | 2.01 ± 0.06 ab | |
| 4012 | 20.47 ± 0.67 a | 2.31 ± 0.09 a | 2.24 ± 0.14 a | |
| 0279 | 16.97 ± 0.85 b | 1.76 ± 0.06 b | 1.81 ± 0.06 b | |
| 3538 | 17.33 ± 0.67 b | 1.64 ± 0.02 b | 1.65 ± 0.03 b | |
| WT | BL | 19.00 ± 0.75 bc | 1.95 ± 0.2 bc | 1.99 ± 0.09 bc |
| 3005 | 44.30 ± 4.84 a | 4.14 ± 0.49 a | 4.09 ± 0.46 a | |
| 4012 | 27.00 ± 1.68 b | 2.57 ± 0.22 b | 2.68 ± 0.22 b | |
| 0279 | 13.60 ± 1.81 c | 1.16 ± 0.11 c | 1.26 ± 0.06 c | |
| 3538 | 21.93 ± 0.43 b | 1.8 ± 0.04 bc | 1.88 ± 0.05 c | |
| WT | GL | 6.00 ± 0.20 c | 0.5 ± 0.03 c | 0.61 ± 0.03 c |
| 3005 | 18.87 ± 0.9 a | 1.78 ± 0.04 a | 2.35 ± 0.12 a | |
| 4012 | 10.05 ± 2.45 b | 0.73 ± 0.08 b | 1.11 ± 0.24 b | |
| 0279 | 5.80 ± 0.76 c | 0.44 ± 0.07 c | 0.45 ± 0.09 c | |
| 3538 | 6.00 ± 0.60 c | 0.39 ± 0.03 c | 0.54 ± 0.06 c |
| Variants | Light | Pn | Tr | DW | Y(II) | NPQ |
|---|---|---|---|---|---|---|
| WT | WFL | 15.8 ± 0.8 a | 0.34 ± 0.03 b | 15.60 ± 0.79 a | 0.32 ± 0.01 c | 1.7 ± 0.10 a |
| 3005 | 10.7 ± 0.7 b | 0.37 ± 0.05 b | 12.18 ± 0.14 b | 0.50 ± 0.01 b | 0.80 ± 0.02 c | |
| 4012 | 13.9 ± 1.2 ab | 0.52 ± 0.03 a | 12.53 ± 1.24 b | 0.52 ± 0.01 b | 0.71 ± 0.04 c | |
| 0279 | 14.4 ± 0.5 ab | 0.36 ± 0.02 b | 12.68 ± 1.41 b | 0.55 ± 0.01 a | 0.74 ± 0.04 c | |
| 3538 | 10.5 ± 0.5 b | 0.29 ± 0.04 c | 13.52 ± 0.41 b | 0.53 ± 0.02 ab | 0.89 ± 0.03 d | |
| WT | WL | 8.4 ± 0.3 b | 0.29 ± 0.01 c | 9.84 ± 0.32 c | 0.27 ± 0.01 c | 0.92 ± 0.08 c |
| 3005 | 8.0 ± 0.2 b | 0.76 ± 0.03 a | 12.09 ± 0.17 a | 0.40 ± 0.01 b | 1.29 ± 0.12 b | |
| 4012 | 13.5 ± 1.2 a | 0.11 ± 0.02 d | 11.46 ± 0.31 b | 0.21 ± 0.02 d | 0.99 ± 0.01 c | |
| 0279 | 10.3 ± 1.0 ab | 0.34 ± 0.02 c | 12.05 ± 0.43 a | 0.46 ± 0.01 a | 0.94 ± 0.09 c | |
| 3538 | 12.1 ± 0.4 a | 0.62 ± 0.04 b | 11.15 ± 0.28 b | 0.41 ± 0.02 b | 1.81 ± 0.07 a | |
| WT | RL | 14.0 ± 0.7 ab | 0.58 ± 0.02 c | 8.81 ± 0.32 c | 0.40 ± 0.04 b | 0.71 ± 0.10 c |
| 3005 | 12.1 ± 0.8 b | 0.39 ± 0.04 d | 11.08 ± 0.33 b | 0.28 ± 0.02 c | 0.61 ± 0.07 c | |
| 4012 | 6.7 ± 0.7 c | 0.38 ± 0.01 d | 9.66 ± 0.29 c | 0.49 ± 0.02 a | 0.84 ± 0.18 b | |
| 0279 | 8.6 ± 0.4 c | 0.73 ± 0.03 b | 11.69 ± 0.30 b | 0.43 ± 0.01 b | 1.38 ± 0.04 a | |
| 3538 | 16.0 ± 0.4 a | 1.19 ± 0.02 a | 13.85 ± 0.15 a | 0.49 ± 0.02 a | 1.25 ± 0.16 a | |
| WT | BL | 12.7 ± 1.0 ab | 0.60 ± 0.05 c | 12.48 ± 0.57 a | 0.52 ± 0.01 a | 1.01 ± 0.03 b |
| 3005 | 9.2 ± 0.8 b | 0.91 ± 0.06 b | 9.72 ± 1.02 bc | 0.55 ± 0.02 a | 0.99 ± 0.15 b | |
| 4012 | 9.6 ± 1.7 ab | 0.11 ± 0.01 d | 8.65 ± 0.47 c | 0.42 ± 0.03 b | 1.51± 0.12 a | |
| 0279 | 11.3 ± 0.9 b | 0.40 ± 0.07 c | 10.29 ± 0.10 b | 0.55 ± 0.03 a | 0.97 ± 0.18 b | |
| 3538 | 16.0 ± 1.1 a | 2.07 ± 0.16 a | 12.09 ± 1.09 a | 0.58 ± 0.02 a | 0.97 ± 0.06 b | |
| WT | GL | 9.1 ± 0.4 b | 0.40 ± 0.03 a | 11.66 ± 0.37 a | 0.45 ± 0.03 b | 0.85 ± 0.03 c |
| 3005 | 7.7 ± 0.9 bc | 0.14 ± 0.03 c | 12.26 ± 0.55 a | 0.50 ± 0.03 a | 1.18 ± 0.13 b | |
| 4012 | 9.5 ± 1.0 ab | 0.17 ± 0.04 c | 12.32 ± 0.67 a | 0.30 ± 0.02 c | 0.81 ± 0.11 c | |
| 0279 | 6.7 ± 0.6 c | 0.19 ± 0.02 c | 10.93 ± 0.38 b | 0.49 ± 0.01 a | 1.17 ± 0.09 b | |
| 3538 | 11.4 ± 0.6 a | 0.26 ± 0.06 b | 11.0 ± 0.51 b | 0.48 ± 0.01 a | 1.61 ± 0.05 a |
| Variants | Light | ADA | ABA | ADA | ABA | ABA |
|---|---|---|---|---|---|---|
| Tr III | Tr III | Epid Cells | St | Epid Cells | ||
| WT | WFL | 4.7 ± 0.1 b | 14 ± 1.6 c | 711.6 ± 54.3 b | 180.7 ± 1.9 c | 944 ± 167.4 ab |
| 3005 | 2.6 ± 0.4 c | 27.7 ± 5 b | 906.6 ± 18.9 a | 176.4 ± 17 c | 529.3 ± 25.1 d | |
| 4012 | 13.6 ± 1.7 a | 26.4 ± 1.1 b | 751 ± 24 b | 137 ± 7.3 d | 1036.5 ± 53.6 a | |
| 0279 | 4.7 ± 0.8 b | 11.1 ± 1.2 c | 990.1 ± 93.4 a | 324.7 ± 20.1 a | 866.8 ± 28.7 b | |
| 3538 | 6 ± 3.1 bc | 43.1 ± 4.1 a | 972.9 ± 56.6 a | 229.5 ± 35.7 b | 728 ± 0.6 c | |
| WT | WL | 17.1 ± 0.2 a | 36.1 ± 4.8 b | 1135.8 ± 31.6 b | 106.3 ± 1.86 e | 1364.6 ± 167.4 b |
| 3005 | 3.4 ± 0.4 d | 6.5 ± 0.9 d | 1016.6 ± 87.9 c | 131.9 ± 5.2 d | 399.3 ± 20.8 d | |
| 4012 | 14.8 ± 1.5 b | 20.2 ± 5 c | 1789.8 ± 152.1 a | 435.4 ± 22.5 a | 2004.9 ± 120 a | |
| 0279 | 11.8 ± 1.6 c | 46 ± 2.9 a | 1609.5 ± 24.1 a | 305.3 ± 7.9 b | 1299 ± 98.8 b | |
| 3538 | 3.2 ± 0.6 d | 31.8 ± 3.8 b | 820.3 ± 34.2 d | 230.1 ± 13.4 c | 956.6 ± 55 c | |
| WT | RL | 44.7 ± 3.2 a | 182.8 ± 14 a | 1383 ± 106.6 b | 339.9 ± 5.3 c | 2091.4 ± 42.6 a |
| 3005 | 2.2 ± 0.3 e | 6.4 ± 0.5 e | 1159.5 ± 64.2 c | 332.1 ± 10.4 c | 981.9 ± 63.6 d | |
| 4012 | 31.8 ± 3.9 b | 122.2 ± 4.8 b | 1432.9 ± 56.2 b | 376.2 ± 2.9 b | 1304.1 ± 4 c | |
| 0279 | 4.7 ± 0.6 d | 82.6 ± 8.2 c | 2422.7 ± 65 a | 476.2 ± 20.8 a | 1729.4 ± 28.5 b | |
| 3538 | 6.8 ± 0.7 c | 38.5 ± 4.1 d | 881.5 ± 41.3 d | 272.1 ± 1.9 d | 888.7 ± 60.7 d | |
| WT | BL | 26.0 ± 2.4 a | 123.6 ± 8.7 a | 653.7 ± 37.2 b | 244.8 ± 5.2 b | 1122.1 ± 40.4 b |
| 3005 | 16.7 ± 2 b | 64.7 ± 3.3 c | 644.8 ± 10.3 b | 217.5 ± 12.3 c | 843.2 ± 15.4 c | |
| 4012 | 13.9 ± 1.9 b | 87.4 ± 2.4 b | 2001.2 ± 103.4 a | 442.3 ± 15.7 a | 1681 ± 97.6 a | |
| 0279 | 17.2 ± 1.3 b | 41.5 ± 1.6 d | 545.1 ± 34.2 c | 267.4 ± 7.3 b | 832.5 ± 4.6 c | |
| 3538 | 7.0 ± 1.1 d | 30.8 ± 2.3 d | 785 ± 40.2 b | 219.6 ± 3.4 c | 705.5 ± 11.2 d | |
| WT | GL | 19.4 ± 2.1 a | 81.9 ± 6.1 b | 1506.6 ± 45.7 b | 306.9 ± 4.5 b | 2049.5 ± 100.3 a |
| 3005 | 3.7 ± 0.4 b | 17.5 ± 0.1 c | 788.4 ± 15.3 c | 170.6 ± 13.9 d | 714.9 ± 26.8 d | |
| 4012 | 4.2 ± 0.3 b | 114.8 ± 10 a | 2319.9 ± 108 a | 394.5 ± 13.3 a | 1634.6 ± 12.5 b | |
| 0279 | 3.3 ± 0.2 b | 22.6 ± 1.4 c | 1468.9 ± 71.2 b | 266.7 ± 14.4 c | 2050.2 ± 97.6 a | |
| 3538 | 4.5 ± 0.3 b | 15 ± 0.4 c | 1567.8 ± 36.9 b | 128.7 ± 8.0 d | 941.8 ± 61.8 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vereshchagin, M.; Kreslavski, V.; Ivanov, Y.; Ivanova, A.; Kumachova, T.; Ryabchenko, A.; Kosobryukhov, A.; Kuznetsov, V.; Pashkovskiy, P. Investigating the Physiological and Molecular Responses of Solanum lycopersicum hp Mutants to Light of Different Quality for Biotechnological Applications. Int. J. Mol. Sci. 2023, 24, 10149. https://doi.org/10.3390/ijms241210149
Vereshchagin M, Kreslavski V, Ivanov Y, Ivanova A, Kumachova T, Ryabchenko A, Kosobryukhov A, Kuznetsov V, Pashkovskiy P. Investigating the Physiological and Molecular Responses of Solanum lycopersicum hp Mutants to Light of Different Quality for Biotechnological Applications. International Journal of Molecular Sciences. 2023; 24(12):10149. https://doi.org/10.3390/ijms241210149
Chicago/Turabian StyleVereshchagin, Mikhail, Vladimir Kreslavski, Yury Ivanov, Alexandra Ivanova, Tamara Kumachova, Andrey Ryabchenko, Anatoliy Kosobryukhov, Vladimir Kuznetsov, and Pavel Pashkovskiy. 2023. "Investigating the Physiological and Molecular Responses of Solanum lycopersicum hp Mutants to Light of Different Quality for Biotechnological Applications" International Journal of Molecular Sciences 24, no. 12: 10149. https://doi.org/10.3390/ijms241210149