Innovative Approach to Isolate and Characterize Glioblastoma Circulating Tumor Cells and Correlation with Tumor Mutational Status
Abstract
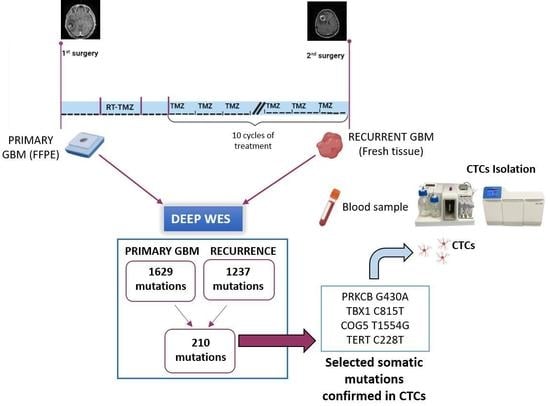
:1. Introduction
2. Results
2.1. CTCs Description
2.2. CNA Analysis on Whole Tissues and CTCs
2.3. Whole Exome Sequencing
2.4. Detection of Patient-Tailored Tumor Somatic Variations in CTCs and BRAF V600E
2.5. Detection of TERT Promoter Variations in CTCs
3. Discussion
4. Materials and Methods
4.1. Patient and Samples
4.2. Study Design
4.3. CTCs Enrichment and Isolation
4.3.1. CTCs Enrichment from Peripheral Whole Blood
4.3.2. Immunofluorescence of Single-Cell Suspensions
4.3.3. Single Cell Isolation by DEPArrayTM NxT
4.3.4. Ampli1™ Whole Genome Amplification and Low-Pass Analysis on CTCs
4.3.5. CNA Calling
4.3.6. DNA Extraction and Whole Exome Sequencing
4.3.7. NGS Analysis
4.3.8. Molecular Characterization of CTCs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, D.; Shen, L.; Luo, M.; Zhang, K.; Li, J.; Yang, Q.; Zhu, F.; Zhou, D.; Zheng, S.; Chen, Y.; et al. Circulating tumor cells: Biology and clinical significance. Signal Transduct. Target. Ther. 2021, 6, 404. [Google Scholar] [CrossRef] [PubMed]
- Sleeman, J.P.; Nazarenko, I.; Thiele, W. Do all roads lead to Rome? Routes to metastasis development. Int. J. Cancer 2011, 128, 2511–2526. [Google Scholar] [CrossRef] [PubMed]
- Vasseur, A.; Kiavue, N.; Bidard, F.C.; Pierga, J.Y.; Cabel, L. Clinical utility of circulating tumor cells: An update. Mol. Oncol. 2021, 15, 1647. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.J.; Halabi, S.; Luo, J.; Nanus, D.M.; Giannakakou, P.; Szmulewitz, R.Z.; Danila, D.C.; Healy, P.; Anand, M.; Rothwell, C.J.; et al. Prospective Multicenter Validation of Androgen Receptor Splice Variant 7 and Hormone Therapy Resistance in High-Risk Castration-Resistant Prostate Cancer: The PROPHECY Study. J. Clin. Oncol. 2019, 37, 1120. [Google Scholar] [CrossRef]
- Cabel, L.; Berger, F.; Cottu, P.; Loirat, D.; Rampanou, A.; Brain, E.; Cyrille, S.; Bourgeois, H.; Kiavue, N.; Deluche, E.; et al. Clinical utility of circulating tumour cell-based monitoring of late-line chemotherapy for metastatic breast cancer: The randomised CirCe01 trial. Br. J. Cancer 2021, 124, 1207. [Google Scholar] [CrossRef]
- Smerage, J.B.; Barlow, W.E.; Hortobagyi, G.N.; Winer, E.P.; Leyland-Jones, B.; Srkalovic, G.; Tejwani, S.; Schott, A.F.; O’Rourke, M.A.; Lew, D.L.; et al. Circulating Tumor Cells and Response to Chemotherapy in Metastatic Breast Cancer: SWOG S0500. J. Clin. Oncol. 2014, 32, 3483. [Google Scholar] [CrossRef]
- Aranda, E.; Viéitez, J.M.; Gómez-España, A.; Calle, S.G.; Salud-Salvia, A.; Graña, B.; Garcia-Alfonso, P.; Rivera, F.; Quintero-Aldana, G.A.; Reina-Zoilo, J.J.; et al. FOLFOXIRI plus bevacizumab versus FOLFOX plus bevacizumab for patients with metastatic colorectal cancer and ≥3 circulating tumour cells: The randomised phase III VISNÚ-1 trial. ESMO Open 2020, 5, e000944. [Google Scholar] [CrossRef]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Hamilton, J.D.; Rapp, M.; Schneiderhan, T.M.; Sabel, M.; Hayman, A.; Scherer, A.; Kröpil, P.; Budach, W.; Kretschmar, U.; Gerber, P.A.; et al. Glioblastoma multiforme metastasis outside the CNS: Three case reports and possible mechanisms of escape. J. Clin. Oncol. 2014, 32, e80–e84. [Google Scholar] [CrossRef]
- Lah, T.T.; Novak, M.; Breznik, B. Brain malignancies: Glioblastoma and brain metastases. Semin. Cancer Biol. 2020, 60, 262–273. [Google Scholar] [CrossRef]
- Pansera, F.; Pansera, E. An explanation for the rarity of extraaxial metastases in brain tumors. Med. Hypotheses 1992, 39, 88–89. [Google Scholar] [CrossRef] [PubMed]
- Jimsheleishvili, S.; Alshareef, A.T.; Papadimitriou, K.; Bregy, A.; Shah, A.H.; Graham, R.M.; Ferraro, N.; Komotar, R.J. Extracranial glioblastoma in transplant recipients. J. Cancer Res. Clin. Oncol. 2014, 140, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.; Holtschmidt, J.; Auer, M.; Heitzer, E.; Lamszus, K.; Schulte, A.; Matschke, J.; Langer-Freitag, S.; Gasch, C.; Stoupiec, M.; et al. Hematogenous dissemination of glioblastoma multiforme. Sci. Transl. Med. 2014, 6, 247ra101. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Cui, Y.; Jiang, H.; Sui, D.; Wang, Y.; Jiang, Z.; Zhao, J.; Lin, S. Circulating tumor cell is a common property of brain glioma and promotes the monitoring system. Oncotarget 2016, 7, 71330–71340. [Google Scholar] [CrossRef] [PubMed]
- Lynch, D.; Powter, B.; Po, J.W.; Cooper, A.; Garrett, C.; Koh, E.S.; Sheridan, M.; van Gelder, J.; Darwish, B.; Mckechnie, S.; et al. Isolation of Circulating Tumor Cells from Glioblastoma Patients by Direct Immunomagnetic Targeting. Appl. Sci. 2020, 10, 3338. [Google Scholar] [CrossRef]
- Sullivan, J.P.; Nahed, B.V.; Madden, M.W.; Oliveira, S.M.; Springer, S.; Bhere, D.; Chi, A.S.; Wakimoto, H.; Michael Rothenberg, S.; Sequist, L.V.; et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014, 4, 1299–1309. [Google Scholar] [CrossRef] [Green Version]
- Krol, I.; Castro-Giner, F.; Maurer, M.; Gkountela, S.; Szczerba, B.M.; Scherrer, R.; Coleman, N.; Carreira, S.; Bachmann, F.; Anderson, S.; et al. Detection of circulating tumour cell clusters in human glioblastoma. Br. J. Cancer 2018, 119, 487–491. [Google Scholar] [CrossRef] [Green Version]
- Gatto, L.; Franceschi, E.; Di Nunno, V.; Tosoni, A.; Lodi, R.; Brandes, A.A. Liquid Biopsy in Glioblastoma Management: From Current Research to Future Perspectives. Oncologist 2021, 26, 865–878. [Google Scholar] [CrossRef]
- MacArthur, K.M.; Kao, G.D.; Chandrasekaran, S.; Alonso-Basanta, M.; Chapman, C.; Lustig, R.A.; Wileyto, E.P.; Hahn, S.M.; Dorsey, J.F. Detection of brain tumor cells in the peripheral blood by a telomerase promoter-based assay. Cancer Res. 2014, 74, 2152–2159. [Google Scholar] [CrossRef] [Green Version]
- Bang-Christensen, S.R.; Pedersen, R.S.; Pereira, M.A.; Clausen, T.M.; Løppke, C.; Sand, N.T.; Ahrens, T.D.; Jørgensen, A.M.; Lim, Y.C.; Goksøyr, L.; et al. Capture and Detection of Circulating Glioma Cells Using the Recombinant VAR2CSA Malaria Protein. Cells 2019, 8, 998. [Google Scholar] [CrossRef] [Green Version]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, M.E. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016, 20, S2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, A.C.; Ashley, D.M.; López, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA. Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Maher, E.A.; Bachoo, R.M. Glioblastoma. In Rosenberg’s Molecular and Genetic Basis of Neurological and Psychiatric Disease, 5th ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 909–917. [Google Scholar] [CrossRef]
- Kim, J.; Lee, I.H.; Cho, H.J.; Park, C.K.; Jung, Y.S.; Kim, Y.; Nam, S.H.; Kim, B.S.; Johnson, M.D.; Kong, D.S.; et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell 2015, 28, 318–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiffer, D.; Giordana, M.T.; Germano, I.; Mauro, A. Anaplasia and heterogeneity of GFAP expression in gliomas. Tumori 1986, 72, 163–170. [Google Scholar] [CrossRef]
- Nel, I.; Jehn, U.; Gauler, T.; Hoffmann, A.C. Individual profiling of circulating tumor cell composition in patients with non-small cell lung cancer receiving platinum based treatment. Transl. Lung Cancer Res. 2014, 3, 100. [Google Scholar] [CrossRef]
- Schneck, H.; Gierke, B.; Uppenkamp, F.; Behrens, B.; Niederacher, D.; Stoecklein, N.H.; Templin, M.F.; Pawlak, M.; Fehm, T.; Neubauer, H. EpCAM-Independent Enrichment of Circulating Tumor Cells in Metastatic Breast Cancer. PLoS ONE 2015, 10, e0144535. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373. [Google Scholar] [CrossRef] [Green Version]
- Lustberg, M.B.; Balasubramanian, P.; Miller, B.; Garcia-Villa, A.; Deighan, C.; Wu, Y.; Carothers, S.; Berger, M.; Ramaswamy, B.; Macrae, E.R.; et al. Heterogeneous atypical cell populations are present in blood of metastatic breast cancer patients. Breast Cancer Res. 2014, 16, R23. [Google Scholar] [CrossRef] [Green Version]
- Khoo, B.L.; Warkiani, M.E.; Tan, D.S.W.; Bhagat, A.A.S.; Irwin, D.; Lau, D.P.; Lim, A.S.T.; Lim, K.H.; Krisna, S.S.; Lim, W.T.; et al. Clinical Validation of an Ultra High-Throughput Spiral Microfluidics for the Detection and Enrichment of Viable Circulating Tumor Cells. PLoS ONE 2014, 9, e99409. [Google Scholar] [CrossRef] [Green Version]
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Michael Rothenberg, S.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gast, C.E.; Silk, A.D.; Zarour, L.; Riegler, L.; Burkhart, J.G.; Gustafson, K.T.; Parappilly, M.S.; Roh-Johnson, M.; Goodman, J.R.; Olson, B.; et al. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci. Adv. 2018, 4, 7828–7840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arita, H.; Narita, Y.; Fukushima, S.; Tateishi, K.; Matsushita, Y.; Yoshida, A.; Miyakita, Y.; Ohno, M.; Collins, V.P.; Kawahara, N.; et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013, 126, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franch-Expósito, S.; Bassaganyas, L.; Vila-Casadesús, M.; Hernández-Illán, E.; Esteban-Fabró, R.; Díaz-Gay, M.; Lozano, J.J.; Castells, A.; Llovet, J.M.; Castellví-Bel, S.; et al. CNApp, a tool for the quantification of copy number alterations and integrative analysis revealing clinical implications. eLife 2020, 9, e50267. [Google Scholar] [CrossRef]
- Guo, Y.; Ding, X.; Shen, Y.; Lyon, G.J.; Wang, K. SeqMule: Automated pipeline for analysis of human exome/genome sequencing data. Sci. Rep. 2015, 5, 14283. [Google Scholar] [CrossRef] [Green Version]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lessi, F.; Morelli, M.; Franceschi, S.; Aretini, P.; Menicagli, M.; Marranci, A.; Pasqualetti, F.; Gambacciani, C.; Pieri, F.; Grimod, G.; et al. Innovative Approach to Isolate and Characterize Glioblastoma Circulating Tumor Cells and Correlation with Tumor Mutational Status. Int. J. Mol. Sci. 2023, 24, 10147. https://doi.org/10.3390/ijms241210147
Lessi F, Morelli M, Franceschi S, Aretini P, Menicagli M, Marranci A, Pasqualetti F, Gambacciani C, Pieri F, Grimod G, et al. Innovative Approach to Isolate and Characterize Glioblastoma Circulating Tumor Cells and Correlation with Tumor Mutational Status. International Journal of Molecular Sciences. 2023; 24(12):10147. https://doi.org/10.3390/ijms241210147
Chicago/Turabian StyleLessi, Francesca, Mariangela Morelli, Sara Franceschi, Paolo Aretini, Michele Menicagli, Andrea Marranci, Francesco Pasqualetti, Carlo Gambacciani, Francesco Pieri, Gianluca Grimod, and et al. 2023. "Innovative Approach to Isolate and Characterize Glioblastoma Circulating Tumor Cells and Correlation with Tumor Mutational Status" International Journal of Molecular Sciences 24, no. 12: 10147. https://doi.org/10.3390/ijms241210147