Enhanced Tumor Imaging Using Glucosamine-Conjugated Polyacrylic Acid-Coated Ultrasmall Gadolinium Oxide Nanoparticles in Magnetic Resonance Imaging
Abstract
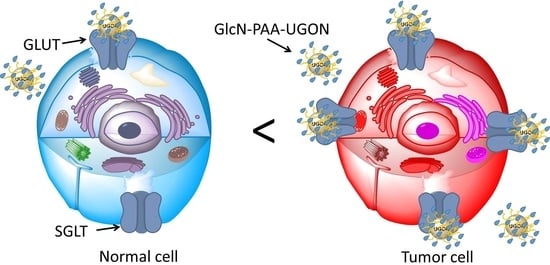
:1. Introduction
2. Results
2.1. Physicochemical Properties of PAA-UGONs and GlcN-PAA-UGONs
2.2. Crystal Structures
2.3. Surface-Coating Results
2.4. Magnetic Properties
2.5. r1 and r2 Values and R1 and R2 Map Images
2.6. In Vitro Cellular Cytotoxicity Results
2.7. In Vivo Tumor Imaging: T1 MR Images in U87MG Tumor-Bearing Mice
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. One-Pot Polyol Synthesis of the PAA-UGONs
4.3. Synthesis of the GlcN-PAA-UGONs
4.4. Physicochemical Property Characterization
4.5. In Vitro Cellular Cytotoxicity Measurements
4.6. Water Proton Spin Relaxivity and Map Image Measurements
4.7. Animal Experiments
4.8. Tumor Model Nude Mice Preparation
4.9. In Vivo T1 MR Image Measurements
5. Conclusions
- (1)
- The particle diameter was ultrasmall (average diameter = 1.76 nm).
- (2)
- The PAA-UGONs and GlcN-PAA-UGONs exhibited excellent colloidal stability in aqueous media (no precipitation after synthesis, >1 year) and low cellular toxicities up to 0.5 mM Gd.
- (3)
- The PAA-UGONs and GlcN-PAA-UGONs exhibited three to four times higher r1 values than those of commercial Gd-chelates, and their r2/r1 ratios were close to one, indicating that they are potential high-performance T1 MRI contrast agents.
- (4)
- The GlcN-PAA-UGONs exhibited higher contrast enhancements at various organs, including the tumor site, compared with the PAA-UGONs, and such contrast enhancements were the highest at the tumor site, owing to the GlcN effects. Consequently, the GlcN-PAA-UGONs can be applied to highly enhancing contrasts in tumor.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ghasemi, M.; Nabipour, I.; Omrani, A.; Alipour, Z.; Assadi, M. Precision medicine and molecular imaging: New targeted approaches toward cancer therapeutic and diagnosis. Am. J. Nucl. Med. Mol. Imaging 2016, 6, 310–327. [Google Scholar] [PubMed]
- Kircher, M.F.; Hricak, H.; Larson, S.M. Molecular imaging for personalized cancer care. Mol. Oncol. 2012, 6, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Haris, M.; Yadav, S.K.; Rizwan, A.; Singh, A.; Wang, E.; Hariharan, H.; Reddy, R.; Marincola, F.M. Molecular magnetic resonance imaging in cancer. J. Transl. Med. 2015, 13, 313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Feng, F.; Jin, Z. Cancer diagnosis and treatment guidance: Role of MRI and MRI probes in the era of molecular imaging. Curr. Pharm. Biotechnol. 2013, 14, 714–722. [Google Scholar] [CrossRef]
- Attia, M.F.; Anton, N.; Wallyn, J.; Omran, Z.; Vandamme, T.F. An overview of active and passive targeting strategies to improve the nanocarriers efficiency to tumor sites. J. Pharm. Pharmacol. 2019, 71, 1185–1198. [Google Scholar] [CrossRef] [Green Version]
- Danhier, F.; Feron, O.; Préat, V. To exploit the tumor microenvironment: Passive and active tumor targeting of nanocarriers for anti-cancer drug delivery. J. Control. Release 2010, 148, 135–146. [Google Scholar] [CrossRef]
- Ahmad, M.Y.; Yue, H.; Tegafaw, T.; Liu, S.; Ho, S.L.; Lee, G.H.; Nam, S.-W.; Chang, Y. Functionalized lanthanide oxide nanoparticles for tumor targeting, medical imaging, and therapy. Pharmaceutics 2021, 13, 1890. [Google Scholar] [CrossRef]
- Tee, J.K.; Yip, L.X.; Tan, E.S.; Santitewagun, S.; Prasath, A.; Ke, P.C.; Ho, H.K.; Leong, D.T. Nanoparticles’ interactions with vasculature in diseases. Chem. Soc. Rev. 2019, 48, 5381–5407. [Google Scholar] [CrossRef]
- Navale, A.M.; Paranjape, A.N. Glucose transporters: Physiological and pathological roles. Biophys. Rev. 2016, 8, 5–9. [Google Scholar] [CrossRef] [Green Version]
- Jang, M.; Kim, S.S.; Lee, J. Cancer cell metabolism: Implications for therapeutic targets. Exp. Mol. Med. 2013, 45, e45. [Google Scholar] [CrossRef] [Green Version]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henrotin, Y.; Mobasheri, A.; Marty, M. Is there any scientific evidence for the use of glucosamine in the management of human osteoarthritis? Arthritis Res. Ther. 2012, 14, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akella, N.M.; Ciraku, L.; Reginato, M.J. Fueling the fire: Emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. 2019, 17, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, J.W.; Nicolosi, R.J.; Borzelleca, J.F. Glucosamine effects in humans: A review of effects on glucose metabolism, side effects, safety considerations and efficacy. Food Chem. Toxicol. 2005, 43, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.R.; Marks, V.; Leeds, A.R.; Anderson, J.W. A comprehensive review of oral glucosamine use and effects on glucose metabolism in normal and diabetic individuals. Diabetes Metab. Res. Rev. 2011, 27, 14–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorens, B.; Mueckler, M. Glucose transporters in the 21st century. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E141–E145. [Google Scholar] [CrossRef] [Green Version]
- Szablewski, L. Expression of glucose transporters in cancers. Biochim. Biophys. Acta 2013, 1835, 164–169. [Google Scholar] [CrossRef]
- Pooja, D.; Reddy, T.S.; Kulhari, H.; Kadari, A.; Adams, D.J.; Bansal, V.; Sistla, R. N-acetyl-D-glucosamine-conjugated PAMAM dendrimers as dual receptor-targeting nanocarriers for anticancer drug delivery. Eur. J. Pharm. Biopharm. 2020, 154, 377–386. [Google Scholar] [CrossRef]
- Ghanbari, N.; Salehi, Z.; Khodadadi, A.A.; Shokrgozar, M.A.; Saboury, A.A. Glucosamine-conjugated graphene quantum dots as versatile and pH-sensitive nanocarriers for enhanced delivery of curcumin targeting to breast cancer. Mater. Sci. Eng. C 2021, 121, 111809. [Google Scholar] [CrossRef]
- Pawar, S.; Vavia, P. Glucosamine anchored cancer targeted nano-vesicular drug delivery system of doxorubicin. J. Drug Target. 2016, 24, 68–79. [Google Scholar] [CrossRef]
- Ranjbar-Navazi, Z.; Eskandani, M.; Johari-Ahar, M.; Nemati, A.; Akbari, H.; Davaran, S.; Omidi, Y. Doxorubicin-conjugated D-glucosamine- and folate-bi-functionalised InP/ZnS quantum dots for cancer cells imaging and therapy. J. Drug Target. 2018, 26, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Greenwood, T.R.; Glunde, K. Glucosamine-bound near-infrared fluorescent probes with lysosomal specificity for breast tumor imaging. Neoplasia 2008, 10, 389–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korotcov, A.V.; Ye, Y.; Chen, Y.; Zhang, F.; Huang, S.; Lin, S.; Sridhar, R.; Achilefu, S.; Wang, P.C. Glucosamine-linked near-infrared fluorescent probes for imaging of solid tumor xenografts. Mol. Imaging Biol. 2012, 14, 443–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patharkar, A.; Raval, N.; Kalyane, D.; Tambe, V.; Anup, N.; More, N.; Kapusetti, G.; Kalia, K.; Tekade, R.K. Glucosamine-conjugated nanoseeds for chemo-magneto hyperthermia therapy of cancer. J. Drug Delv. Sci. Technol. 2021, 61, 102295. [Google Scholar] [CrossRef]
- Nagura, K.; Takemoto, Y.; Yoshino, F.; Bogdanov, A.; Chumakova, N.; Vorobiev, A.K.; Imai, H.; Matsuda, T.; Shimono, S.; Kato, T.; et al. Magnetic mixed micelles composed of a non-ionic surfactant and nitroxide radicals containing a D-glucosamine unit: Preparation, stability, and biomedical application. Pharmaceutics 2019, 11, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortezazadeh, T.; Gholibegloo, E.; Riyahi, A.N.; Haghgoo, S.; Musa, A.E.; Khoobi, M. Glucosamine conjugated gadolinium (III) oxide nanoparticles as a novel targeted contrast agent for cancer diagnosis in MRI. J. Biomed. Phys. Eng. 2020, 10, 25–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehravi, B.; Ahmadi, M.; Amanlou, M.; Mostaar, A.; Ardestani, M.S.; Ghalandarlaki, N. Conjugation of glucosamine with Gd3+-based nanoporous silica using a heterobifunctional ANB-NOS crosslinker for imaging of cancer cells. Int. J. Nanomed. 2013, 8, 3383–3394. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Chen, Y.; Guo, D.J.; Huang, Z.W.; Cai, L.; He, L. The synthesis of a D-glucosamine contrast agent, Gd-DTPA-DG, and its application in cancer molecular imaging with MRI. Eur. J. Radiol. 2011, 79, 369–374. [Google Scholar] [CrossRef]
- Wahsner, J.; Gale, E.M.; Rodríguez-Rodríguez, A.; Caravan, P. Chemistry of MRI contrast agents: Current challenges and new frontiers. Chem. Rev. 2018, 119, 957–1057. [Google Scholar] [CrossRef]
- Ho, S.L.; Cha, H.; Oh, I.T.; Jung, K.-H.; Kim, M.H.; Lee, Y.J.; Miao, X.; Tegafaw, T.; Ahmad, M.Y.; Chae, K.S.; et al. Magnetic resonance imaging, gadolinium neutron capture therapy, and tumor cell detection using ultrasmall Gd2O3 nanoparticles coated with polyacrylic acid-rhodamine B as a multifunctional tumor theragnostic agent. RSC Adv. 2018, 8, 12653–12665. [Google Scholar] [CrossRef] [Green Version]
- Jang, Y.J.; Liu, S.; Yue, H.; Park, J.A.; Cha, H.; Ho, S.L.; Marasini, S.; Ghazanfari, A.; Ahmad, M.Y.; Miao, X.; et al. Hydrophilic biocompatible poly(acrylic acid-co-maleic acid) polymer as a surface-coating ligand of ultrasmall Gd2O3 nanoparticles to obtain a high r1 value and T1 MR images. Diagnostics 2021, 11, 2. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.Y.; Ahmad, W.; Yue, H.; Ho, S.L.; Park, J.A.; Jung, K.-H.; Cha, H.; Marasini, S.; Ghazanfari, A.; Liu, S.; et al. In vivo positive magnetic resonance imaging applications of poly(methyl vinyl ether-alt-maleic acid)-coated ultra-small paramagnetic gadolinium oxide nanoparticles. Molecules 2020, 25, 1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, H.; Marasini, S.; Ahmad, M.Y.; Ho, S.L.; Cha, H.; Liu, S.; Jang, Y.J.; Tegafaw, T.; Ghazanfari, A.; Miao, X.; et al. Carbon-coated ultrasmall gadolinium oxide (Gd2O3@C) nanoparticles: Application to magnetic resonance imaging and fluorescence properties. Colloids Surfaces A Physicochem. Eng. Asp. 2019, 586, 124261. [Google Scholar] [CrossRef]
- Park, J.Y.; Baek, M.J.; Choi, E.S.; Woo, S.; Kim, J.H.; Kim, T.J.; Jung, J.C.; Chae, K.S.; Chang, Y.; Lee, G.H. Paramagnetic ultrasmall gadolinium oxide nanoparticles as advanced T1 MRI contrast agent: Account for large longitudinal relaxivity, optimal particle diameter, and in vivo T1 MR images. ACS Nano 2009, 3, 3663–3669. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Liu, W.; Misra, P.; Tanaka, E.; Zimmer, J.P.; Ipe, B.I.; Bawendi, M.G.; Frangioni, J.V. Renal clearance of nano-particles. Nat. Biotechnol. 2007, 25, 1165–1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longmire, M.; Choyke, P.L.; Kobayashi, H. Clearance properties of nano-sized particles and molecules as imaging agents: Con-siderations and caveats. Nanomedicine 2008, 3, 703–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marasini, S.; Yue, H.; Ho, S.L.; Jung, K.-H.; Park, J.A.; Cha, H.; Ghazanfari, A.; Ahmad, M.Y.; Liu, S.; Jang, Y.J.; et al. D-glucuronic acid-coated ultrasmall paramagnetic Ln2O3 (Ln = Tb, Dy, and Ho) nanoparticles: Magnetic properties, water proton relaxivities, and fluorescence properties. Eur. J. Inorg. Chem. 2019, 34, 3822–3833. [Google Scholar] [CrossRef]
- Yim, E.S.; Zhao, B.; Myung, D.; Kourtis, L.C.; Frank, C.W.; Carter, D.; Smith, R.L.; Goodman, S.B. Biocompatibility of poly(ethylene glycol)/poly(acrylic acid) interpenetrating polymer network hydrogel particles in RAW 264.7 macrophage and MG-63 osteoblast cell lines. J. Biomed. Mater. Res. 2009, 91, 894–902. [Google Scholar] [CrossRef]
- Marasini, S.; Yue, H.; Ho, S.L.; Park, J.A.; Kim, S.; Jung, K.-H.; Cha, H.; Liu, S.; Tegafaw, T.; Ahmad, M.Y.; et al. Synthesis, characterization, and 9.4 tesla T2 MR images of polyacrylic acid-coated terbium(III) and holmium(III) oxide nanoparticles. Nanomaterials 2021, 11, 1355. [Google Scholar] [CrossRef]
- Card No. 43-1014, JCPDS-International Centre for Diffraction Data; Version 1.30; PCPDFWIN: Newtown Square, PA, USA, 1997.
- Pearson, R.G. Hard and Soft Acids and Bases. J. Am. Chem. Soc. 1963, 85, 3533–3539. [Google Scholar] [CrossRef]
- Pearson, R.G. Hard and soft acids and bases, HSAB, Part I: Fundamental principles. J. Chem. Educ. 1968, 45, 581–587. [Google Scholar] [CrossRef]
- Pearson, R.G. Hard and soft acids and bases, HSAB, Part II: Underlying theories. J. Chem. Educ. 1968, 45, 643–648. [Google Scholar] [CrossRef]
- Deacon, G.B.; Phillips, R.J. Relationships between the carbon-oxygen stretching frequencies of carboxylato complexes and the type of carboxylate coordination. Coord. Chem. Rev. 1980, 33, 227–250. [Google Scholar] [CrossRef]
- Hao, X.P.; Chen, S.G.; Wang, W.H.; Yang, Z.Q.; Yue, L.F.; Sun, H.Y.; Cheng, F. AgNP-coordinated glucosamine-grafted carbon nanotubes with enhanced antibacterial properties. New J. Chem. 2017, 41, 7045–7051. [Google Scholar] [CrossRef]
- Telange, D.R.; Bhagat, S.B.; Patil, A.T.; Umekar, M.J.; Pethe, A.M.; Raut, N.A.; Dave, V.S. Glucosamine HCl-based solid dispersions to enhance the biopharmaceutical properties of acyclovir. J. Excip. Food Chem. 2019, 10, 65–81. [Google Scholar]
- Zaeni, A.; Safitri, E.; Fuadah, B.; Sudiana, I.N. Microwave-assisted hydrolysis of chitosan from shrimp shell waste for glucosammine hydrochlorid production. IOP Conf. Series J. Phys. Conf. Ser. 2017, 846, 012011. [Google Scholar] [CrossRef] [Green Version]
- Corbierre, M.K.; Cameron, N.S.; Lennox, R.B. Polymer-stabilized gold nanoparticles with high grafting densities. Langmuir 2004, 20, 2867–2873. [Google Scholar] [CrossRef]
- Haynes, W.M.; Lide, D.R.; Bruno, T.J. CRC Handbook of Chemistry and Physics, 96th ed.; CRC Press: Boca Raton, FL, USA, 2015–2016; pp. 4–64. [CrossRef]
- Arajs, S.; Colvin, R.V. Magnetic susceptibility of gadolinium and dysprosium sesquioxides at elevated temperatures. J. Appl. Phys. 1962, 33, 2517–2519. [Google Scholar] [CrossRef]
- Moon, R.M.; Koehler, W.C. Magnetic properties of Gd2O3. Phys. Rev. B 1975, 11, 1609–1622. [Google Scholar] [CrossRef]
- Roch, A.; Muller, R.N.; Gillis, P. Theory of proton relaxation induced by superparamagnetic particles. J. Chem. Phys. 1999, 110, 5403–5411. [Google Scholar] [CrossRef]
- Roch, A.; Gossuin, Y.; Muller, R.N.; Gillis, P. Superparamagnetic colloid suspensions: Water magnetic relaxation and clustering. J. Magn. Magn. Mater. 2005, 293, 532–539. [Google Scholar] [CrossRef]
- Miao, X.; Ho, S.L.; Tegafaw, T.; Cha, H.; Chang, Y.; Oh, I.T.; Ahmad, M.Y.; Marasini, S.; Ghazanfari, A.; Yue, H.; et al. Stable and non-toxic ultrasmall gadolinium oxide nanoparticle colloids (coating material = polyacrylic acid) as high-performance T1 magnetic resonance imaging contrast agents. RSC. Adv. 2018, 8, 3189–3197. [Google Scholar] [CrossRef] [Green Version]
- Akhtar, M.J.; Ahamed, M.; Alhadlaq, H. Gadolinium oxide nanoparticles induce toxicity in human endothelial HUVECs via lipid peroxidation, mitochondrial dysfunction and autophagy modulation. Nanomaterials 2020, 10, 1675. [Google Scholar] [CrossRef] [PubMed]
- Tseng, S.-H.; Lin, S.-M.; Chen, J.-C.; Su, Y.-H.; Huang, H.-Y.; Chen, C.-K.; Lin, P.-Y.; Chen, Y. Resveratrol suppresses the angiogenesis and tumor growth of gliomas in rats. Clin. Cancer Res. 2004, 10, 2190–2202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bäuerle, T.; Komljenovic, D.; Berger, M.R.; Semmler, W. Multi-modal imaging of angiogenesis in a nude rat model of breast cancer bone metastasis using magnetic resonance imaging, volumetric computed tomography and ultrasound. J. Vis. Exp. 2012, 66, e4178. [Google Scholar] [CrossRef] [PubMed] [Green Version]















| Ligand | davg (nm) | aavg (nm) | ξ (mV) | Surface-Coating Results | |||
|---|---|---|---|---|---|---|---|
| P a (wt.%) | UGON (wt.%) | σ b (l/nm2) | NNP c | ||||
| PAA | 1.76 ± 0.01 | 9.2 ± 0.1 | –36.0 ± 0.5 | 41.7 (48.2) | 47.1 | 0.64 | ~6 PAA |
| GlcN-PAA | 1.76 ± 0.01 | 10.6 ± 0.1 | –30.7 ± 0.2 | 50.3 (54.4) | 39.3 | 0.64 | ~6 (PAA + ~5 GlcN) |
| Vibration a | PAA | GlcN | PAA-UGON | GlcN-PAA-UGON | Ref |
|---|---|---|---|---|---|
| (C-O) s | - | 1026 | - | 1060 | [30,45,46] |
| (C-N) s | - | 1415 | - | 1401 | [45] |
| (COO-) ss | - | - | 1401 | 1401 | [30,45] |
| (COO-) as | - | - | 1538 | 1538 | [30,45] |
| (N-H) b | - | 1540 | - | 1538 | [46] |
| (C=O) s | 1700 | - | - | - | [30] |
| (C-H) s | 2957 | 2940 | 2945 | 2945 | [30,45,46] |
| (N-H) s | - | 3283 | - | ~3319 | [45,46,47] |
| (O-H) s | - | 3346 | ~3319 | ~3319 | [45,46,47] |
| Nanoparticle | Magnetic Properties | Water Proton Spin Relaxivities (s−1mM−1) | ||||||
|---|---|---|---|---|---|---|---|---|
| Magnetism | Net M at 2.0 T (emu/g) | H = 1.5 T | H = 3.0 T | |||||
| r1 | r2 | r2/r1 | r1 | r2 | r2/r1 | |||
| PAA-UGON | Paramagnetic | 1.89 | 14.31 | 24.09 | 1.68 | 19.32 | 34.23 | 1.77 |
| GlcN-PAA-UGON | Paramagnetic | 1.98 | 10.18 | 18.38 | 1.80 | 12.60 | 24.46 | 1.92 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Yue, H.; Ho, S.L.; Kim, S.; Park, J.A.; Tegafaw, T.; Ahmad, M.Y.; Kim, S.; Saidi, A.K.A.A.; Zhao, D.; et al. Enhanced Tumor Imaging Using Glucosamine-Conjugated Polyacrylic Acid-Coated Ultrasmall Gadolinium Oxide Nanoparticles in Magnetic Resonance Imaging. Int. J. Mol. Sci. 2022, 23, 1792. https://doi.org/10.3390/ijms23031792
Liu S, Yue H, Ho SL, Kim S, Park JA, Tegafaw T, Ahmad MY, Kim S, Saidi AKAA, Zhao D, et al. Enhanced Tumor Imaging Using Glucosamine-Conjugated Polyacrylic Acid-Coated Ultrasmall Gadolinium Oxide Nanoparticles in Magnetic Resonance Imaging. International Journal of Molecular Sciences. 2022; 23(3):1792. https://doi.org/10.3390/ijms23031792
Chicago/Turabian StyleLiu, Shuwen, Huan Yue, Son Long Ho, Soyeon Kim, Ji Ae Park, Tirusew Tegafaw, Mohammad Yaseen Ahmad, Seungho Kim, Abdullah Khamis Ali Al Saidi, Dejun Zhao, and et al. 2022. "Enhanced Tumor Imaging Using Glucosamine-Conjugated Polyacrylic Acid-Coated Ultrasmall Gadolinium Oxide Nanoparticles in Magnetic Resonance Imaging" International Journal of Molecular Sciences 23, no. 3: 1792. https://doi.org/10.3390/ijms23031792