Macrophage Migration Inhibitory Factor in Major Depressive Disorder: A Multilevel Pilot Study
Abstract
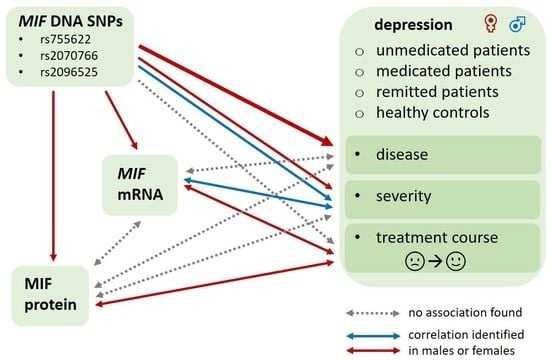
:1. Introduction
2. Results
2.1. Cohort Characteristics
2.2. Influence of MIF Polymorphisms on Risk for and Severity of Depression
2.3. Association of Depression Diagnosis and Severity with MIF Protein and MIF Expression
2.4. Prediction of Treatment Course from MIF Protein and MIF Expression Levels at Inclusion
2.5. Associations of the Treatment Course with Changes of MIF Protein and MIF Expression
2.6. Association between MIF Parameters at Different Biological Levels
3. Discussion
3.1. Influence of MIF Polymorphisms on Risk for and Severity of Depression
3.2. Association of Depression Diagnosis and Severity with MIF Protein and MIF Expression
3.3. Prediction of Treatment Course from Baseline MIF Protein and MIF Expression Levels
3.4. Associations of the Treatment Course with Changes of MIF Protein and MIF Expression
3.5. Association between MIF Parameters at Different Biological Levels
3.6. Strengths and Limitations
4. Materials and Methods
4.1. Study Description
4.2. Psychometric Scales
4.3. Blood Collection and Analysis
4.4. Genotyping MIF Single Nucleotide Polymorphisms
4.5. Quantitative PCR for MIF Expression Analysis
4.6. Enzyme Linked Immunosorbent Assay for MIF Serum Levels
4.7. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Burden of Disease Study. Available online: https://vizhub.healthdata.org/gbd-results/ (accessed on 14 May 2022).
- Young, J.J.; Silber, T.; Bruno, D.; Galatzer-Levy, I.R.; Pomara, N.; Marmar, C.R. Is there Progress? An Overview of Selecting Biomarker Candidates for Major Depressive Disorder. Front. Psychiatry 2016, 7, 72. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, P.F.; Neale, M.C.; Kendler, K.S. Genetic epidemiology of major depression: Review and meta-analysis. Am. J. Psychiatry 2000, 157, 1552–1562. [Google Scholar] [CrossRef]
- Pitsillou, E.; Bresnehan, S.M.; Kagarakis, E.A.; Wijoyo, S.J.; Liang, J.; Hung, A.; Karagiannis, T.C. The cellular and molecular basis of major depressive disorder: Towards a unified model for understanding clinical depression. Mol. Biol. Rep. 2020, 47, 753–770. [Google Scholar] [CrossRef]
- Filatova, E.V.; Shadrina, M.I.; Slominsky, P.A. Major Depression: One Brain, One Disease, One Set of Intertwined Processes. Cells 2021, 10, 1283. [Google Scholar] [CrossRef]
- Schaller, G.; Sperling, W.; Richter-Schmidinger, T.; Muhle, C.; Heberlein, A.; Maihofner, C.; Kornhuber, J.; Lenz, B. Serial repetitive transcranial magnetic stimulation (rTMS) decreases BDNF serum levels in healthy male volunteers. J. Neural Transm. 2014, 121, 307–313. [Google Scholar] [CrossRef]
- Muhle, C.; Bilbao Canalejas, R.D.; Kornhuber, J. Sphingomyelin Synthases in Neuropsychiatric Health and Disease. Neurosignals 2019, 27, 54–76. [Google Scholar] [CrossRef]
- Schumacher, F.; Edwards, M.J.; Muhle, C.; Carpinteiro, A.; Wilson, G.C.; Wilker, B.; Soddemann, M.; Keitsch, S.; Scherbaum, N.; Muller, B.W.; et al. Ceramide levels in blood plasma correlate with major depressive disorder severity and its neutralization abrogates depressive behavior in mice. J. Biol. Chem. 2022, 298, 102185. [Google Scholar] [CrossRef]
- Rhein, C.; Zoicas, I.; Marx, L.M.; Zeitler, S.; Hepp, T.; von Zimmermann, C.; Muhle, C.; Richter-Schmidinger, T.; Lenz, B.; Erim, Y.; et al. mRNA Expression of SMPD1 Encoding Acid Sphingomyelinase Decreases upon Antidepressant Treatment. Int. J. Mol. Sci. 2021, 22, 5700. [Google Scholar] [CrossRef]
- Zoicas, I.; Muhle, C.; Schmidtner, A.K.; Gulbins, E.; Neumann, I.D.; Kornhuber, J. Anxiety and Depression Are Related to Higher Activity of Sphingolipid Metabolizing Enzymes in the Rat Brain. Cells 2020, 9, 1239. [Google Scholar] [CrossRef]
- Muller, C.P.; Reichel, M.; Muhle, C.; Rhein, C.; Gulbins, E.; Kornhuber, J. Brain membrane lipids in major depression and anxiety disorders. Biochim. Biophys. Acta 2015, 1851, 1052–1065. [Google Scholar] [CrossRef]
- Buchholz, V.N.; Muhle, C.; Cohort Study on Substance Use Risk, F.; Kornhuber, J.; Lenz, B. Lower Digit Ratio (2D:4D) Indicative of Excess Prenatal Androgen Is Associated With Increased Sociability and Greater Social Capital. Front. Behav. Neurosci. 2019, 13, 246. [Google Scholar] [CrossRef] [PubMed]
- Lenz, B.; Rother, M.; Bouna-Pyrrou, P.; Muhle, C.; Tektas, O.Y.; Kornhuber, J. The androgen model of suicide completion. Prog. Neurobiol. 2019, 172, 84–103. [Google Scholar] [CrossRef] [PubMed]
- Lenz, B.; Thiem, D.; Bouna-Pyrrou, P.; Muhle, C.; Stoessel, C.; Betz, P.; Kornhuber, J. Low digit ratio (2D:4D) in male suicide victims. J. Neural Transm. 2016, 123, 1499–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenz, B.; Eichler, A.; Schwenke, E.; Buchholz, V.N.; Hartwig, C.; Moll, G.H.; Reich, K.; Muhle, C.; Volz, B.; Titzmann, A.; et al. Mindfulness-based Stress Reduction in Pregnancy: An App-Based Programme to Improve the Health of Mothers and Children (MINDFUL/PMI Study). Geburtshilfe Frauenheilkd 2018, 78, 1283–1291. [Google Scholar] [CrossRef] [Green Version]
- Dantzer, R.; O’Connor, J.C.; Freund, G.G.; Johnson, R.W.; Kelley, K.W. From inflammation to sickness and depression: When the immune system subjugates the brain. Nat. Rev. Neurosci. 2008, 9, 46–56. [Google Scholar] [CrossRef] [Green Version]
- Valentine, A.D.; Meyers, C.A. Neurobehavioral effects of interferon therapy. Curr. Psychiatry Rep. 2005, 7, 391–395. [Google Scholar] [CrossRef]
- Musselman, D.L.; Lawson, D.H.; Gumnick, J.F.; Manatunga, A.K.; Penna, S.; Goodkin, R.S.; Greiner, K.; Nemeroff, C.B.; Miller, A.H. Paroxetine for the prevention of depression induced by high-dose interferon alfa. N. Engl. J. Med. 2001, 344, 961–966. [Google Scholar] [CrossRef]
- Dobos, N.; Vries, E.F.J.; Kema, I.P.; Patas, K.; Prins, M.; Nijholt, I.M.; Dierckx, R.A.; Korf, J.; den Boer, J.A.; Luiten, P.G.M.; et al. The role of indoleamine 2,3-dioxygenase in a mouse model of neuroinflammation-induced depression. J. Alzheimer’s Dis. JAD 2012, 28, 905–915. [Google Scholar] [CrossRef] [Green Version]
- Petralia, M.C.; Mazzon, E.; Fagone, P.; Basile, M.S.; Lenzo, V.; Quattropani, M.C.; Bendtzen, K.; Nicoletti, F. Pathogenic contribution of the Macrophage migration inhibitory factor family to major depressive disorder and emerging tailored therapeutic approaches. J. Affect. Disord. 2020, 263, 15–24. [Google Scholar] [CrossRef]
- Günther, S.; Fagone, P.; Jalce, G.; Atanasov, A.G.; Guignabert, C.; Nicoletti, F. Role of MIF and D-DT in immune-inflammatory, autoimmune, and chronic respiratory diseases: From pathogenic factors to therapeutic targets. Drug Discov. Today 2019, 24, 428–439. [Google Scholar] [CrossRef]
- Calandra, T.; Roger, T. Macrophage migration inhibitory factor: A regulator of innate immunity. Nat. Rev. Immunol. 2003, 3, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Calandra, T.; Bernhagen, J.; Mitchell, R.A.; Bucala, R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J. Exp. Med. 1994, 179, 1895–1902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calandra, T.; Bernhagen, J.; Metz, C.N.; Spiegel, L.A.; Bacher, M.; Donnelly, T.; Cerami, A.; Bucala, R. MIF as a glucocorticoid-induced modulator of cytokine production. Nature 1995, 377, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Renner, P.; Roger, T.; Calandra, T. Macrophage migration inhibitory factor: Gene polymorphisms and susceptibility to inflammatory diseases. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2005, 41 (Suppl. 7), S513–S519. [Google Scholar] [CrossRef] [PubMed]
- Benedek, G.; Meza-Romero, R.; Jordan, K.; Zhang, Y.; Nguyen, H.; Kent, G.; Li, J.; Siu, E.; Frazer, J.; Piecychna, M.; et al. MIF and D-DT are potential disease severity modifiers in male MS subjects. Proc. Natl. Acad. Sci.USA 2017, 114, E8421–E8429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petralia, M.C.; Battaglia, G.; Bruno, V.; Pennisi, M.; Mangano, K.; Lombardo, S.D.; Fagone, P.; Cavalli, E.; Saraceno, A.; Nicoletti, F.; et al. The Role of Macrophage Migration Inhibitory Factor in Alzheimer’s Disease: Conventionally Pathogenetic or Unconventionally Protective? Molecules 2020, 25, 291. [Google Scholar] [CrossRef] [Green Version]
- Bloom, J.; Al-Abed, Y. MIF: Mood improving/inhibiting factor? J. Neuroinflamm. 2014, 11, 11. [Google Scholar] [CrossRef] [Green Version]
- Musil, R.; Schwarz, M.J.; Riedel, M.; Dehning, S.; Cerovecki, A.; Spellmann, I.; Arolt, V.; Müller, N. Elevated macrophage migration inhibitory factor and decreased transforming growth factor-beta levels in major depression—No influence of celecoxib treatment. J. Affect. Disord. 2011, 134, 217–225. [Google Scholar] [CrossRef]
- Edwards, K.M.; Bosch, J.A.; Engeland, C.G.; Cacioppo, J.T.; Marucha, P.T. Elevated macrophage migration inhibitory factor (MIF) is associated with depressive symptoms, blunted cortisol reactivity to acute stress, and lowered morning cortisol. Brain Behav. Immun. 2010, 24, 1202–1208. [Google Scholar] [CrossRef]
- Bay-Richter, C.; Janelidze, S.; Sauro, A.; Bucala, R.; Lipton, J.; Deierborg, T.; Brundin, L. Behavioural and neurobiological consequences of macrophage migration inhibitory factor gene deletion in mice. J. Neuroinflamm. 2015, 12, 163. [Google Scholar] [CrossRef]
- Moon, H.Y.; Kim, S.H.; Yang, Y.R.; Song, P.; Yu, H.S.; Park, H.G.; Hwang, O.; Lee-Kwon, W.; Seo, J.K.; Hwang, D.; et al. Macrophage migration inhibitory factor mediates the antidepressant actions of voluntary exercise. Proc. Natl. Acad. Sci. USA 2012, 109, 13094–13099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conboy, L.; Varea, E.; Castro, J.E.; Sakouhi-Ouertatani, H.; Calandra, T.; Lashuel, H.A.; Sandi, C. Macrophage migration inhibitory factor is critically involved in basal and fluoxetine-stimulated adult hippocampal cell proliferation and in anxiety, depression, and memory-related behaviors. Mol. Psychiatry 2011, 16, 533–547. [Google Scholar] [CrossRef] [PubMed]
- Leyton-Jaimes, M.F.; Kahn, J.; Israelson, A. Macrophage migration inhibitory factor: A multifaceted cytokine implicated in multiple neurological diseases. Exp. Neurol. 2018, 301, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, A.; Gennarelli, M.; Uher, R.; Breen, G.; Farmer, A.; Aitchison, K.J.; Craig, I.W.; Anacker, C.; Zunsztain, P.A.; McGuffin, P.; et al. Candidate genes expression profile associated with antidepressants response in the GENDEP study: Differentiating between baseline ‘predictors’ and longitudinal ‘targets’. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2013, 38, 377–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cattaneo, A.; Ferrari, C.; Uher, R.; Bocchio-Chiavetto, L.; Riva, M.A.; Pariante, C.M. Absolute Measurements of Macrophage Migration Inhibitory Factor and Interleukin-1-β mRNA Levels Accurately Predict Treatment Response in Depressed Patients. Int. J. Neuropsychopharmacol. 2016, 19, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamidi, A.K.; Arzaghi, S.M.; Qorbani, M.; Khatami, F.; Ebrahimi, M.; Bandarian, F.; Enayati, S.; Amoli, M.M. MIF 173 GC variation was associated with depressive disorder in type 2 diabetes in an Iranian population. Psychoneuroendocrinology 2019, 104, 243–248. [Google Scholar] [CrossRef]
- Sparkes, A.; De Baetselier, P.; Brys, L.; Cabrito, I.; Sterckx, Y.G.; Schoonooghe, S.; Muyldermans, S.; Raes, G.; Bucala, R.; Vanlandschoot, P.; et al. Novel half-life extended anti-MIF nanobodies protect against endotoxic shock. FASEB J. 2018, 32, 3411–3422. [Google Scholar] [CrossRef] [Green Version]
- Donn, R.; Alourfi, Z.; Benedetti, F.; Meazza, C.; Zeggini, E.; Lunt, M.; Stevens, A.; Shelley, E.; Lamb, R.; Ollier, W.E.R.; et al. Mutation screening of the macrophage migration inhibitory factor gene: Positive association of a functional polymorphism of macrophage migration inhibitory factor with juvenile idiopathic arthritis. Arthritis Rheum. 2002, 46, 2402–2409. [Google Scholar] [CrossRef]
- Donn, R.; Alourfi, Z.; Zeggini, E.; Lamb, R.; Jury, F.; Lunt, M.; Meazza, C.; Benedetti, F.; Thomson, W.; Ray, D. A functional promoter haplotype of macrophage migration inhibitory factor is linked and associated with juvenile idiopathic arthritis. Arthritis Rheum. 2004, 50, 1604–1610. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, D.; Hou, S.; Zhang, C.; Lei, B.; Xiao, X.; Kijlstra, A.; Yang, P. Association of macrophage migration inhibitory factor gene polymorphisms with Behçet’s disease in a Han Chinese population. Ophthalmology 2012, 119, 2514–2518. [Google Scholar] [CrossRef]
- Shimmyo, N.; Hishimoto, A.; Otsuka, I.; Okazaki, S.; Boku, S.; Mouri, K.; Horai, T.; Takahashi, M.; Ueno, Y.; Shirakawa, O.; et al. Association study of MIF promoter polymorphisms with suicide completers in the Japanese population. Neuropsychiatr. Dis. Treat. 2017, 13, 899–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pain, O.; Hodgson, K.; Trubetskoy, V.; Ripke, S.; Marshe, V.S.; Adams, M.J.; Byrne, E.M.; Campos, A.I.; Carrillo-Roa, T.; Cattaneo, A.; et al. Identifying the Common Genetic Basis of Antidepressant Response. Biol. Psychiatry Glob. Open Sci. 2022, 2, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Wang, X.-Y.; Thapa, S.; Tao, L.-Y.; Wu, S.-Z.; Luo, G.-J.; Wang, L.-P.; Wang, J.-N.; Wang, J.; Li, J.; et al. Macrophage migration inhibitory factor promoter polymorphisms (-794 CATT5-8): Relationship with soluble MIF levels in coronary atherosclerotic disease subjects. BMC Cardiovasc. Disord. 2017, 17, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsuura, S.; Kamezaki, Y.; Yamagishi, N.; Kuwano, Y.; Nishida, K.; Masuda, K.; Tanahashi, T.; Kawai, T.; Arisawa, K.; Rokutan, K. Circulating vascular endothelial growth factor is independently and negatively associated with trait anxiety and depressive mood in healthy Japanese university students. Int. J. Psychophysiol. Off. J. Int. Organ. Psychophysiol. 2011, 81, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Goldman, E. Cytokine Tied to Depression in Pregnant Women. Available online: https://www.mdedge.com/familymedicine/article/25580/womens-health/cytokine-tied-depression-pregnant-women-macrophage (accessed on 5 May 2022).
- Christian, L.M.; Franco, A.; Iams, J.D.; Sheridan, J.; Glaser, R. Depressive symptoms predict exaggerated inflammatory responses to an in vivo immune challenge among pregnant women. Brain Behav. Immun. 2010, 24, 49–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, T.; Pu, S.; Ni, Y.; Gao, M.; Li, X.; Zeng, X. Elevated plasma macrophage migration inhibitor factor as a risk factor for the development of post-stroke depression in ischemic stroke. J. Neuroimmunol. 2018, 320, 58–63. [Google Scholar] [CrossRef]
- Sedlinska, T.; Muhle, C.; Richter-Schmidinger, T.; Weinland, C.; Kornhuber, J.; Lenz, B. Male depression syndrome is characterized by pronounced Cluster B personality traits. J Affect Disord 2021, 292, 725–732. [Google Scholar] [CrossRef]
- Ma, L.; Xu, Y.; Wang, G.; Li, R. What do we know about sex differences in depression: A review of animal models and potential mechanisms. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 89, 48–56. [Google Scholar] [CrossRef]
- Seney, M.L.; Huo, Z.; Cahill, K.; French, L.; Puralewski, R.; Zhang, J.; Logan, R.W.; Tseng, G.; Lewis, D.A.; Sibille, E. Opposite Molecular Signatures of Depression in Men and Women. Biol. Psychiatry 2018, 84, 18–27. [Google Scholar] [CrossRef]
- Arteaga-Henríquez, G.; Simon, M.S.; Burger, B.; Weidinger, E.; Wijkhuijs, A.; Arolt, V.; Birkenhager, T.K.; Musil, R.; Müller, N.; Drexhage, H.A. Low-Grade Inflammation as a Predictor of Antidepressant and Anti-Inflammatory Therapy Response in MDD Patients: A Systematic Review of the Literature in Combination With an Analysis of Experimental Data Collected in the EU-MOODINFLAME Consortium. Front. Psychiatry 2019, 10, 458. [Google Scholar] [CrossRef]
- Simon, M.S.; Burger, B.; Weidinger, E.; Arteaga-Henríquez, G.; Zill, P.; Musil, R.; Drexhage, H.A.; Müller, N. Efficacy of Sertraline Plus Placebo or Add-On Celecoxib in Major Depressive Disorder: Macrophage Migration Inhibitory Factor as a Promising Biomarker for Remission After Sertraline-Results From a Randomized Controlled Clinical Trial. Front. Psychiatry 2021, 12, 615261. [Google Scholar] [CrossRef] [PubMed]
- Sundquist, J.; Palmer, K.; Memon, A.A.; Wang, X.; Johansson, L.M.; Sundquist, K. Long-term improvements after mindfulness-based group therapy of depression, anxiety and stress and adjustment disorders: A randomized controlled trial. Early Interv. Psychiatry 2019, 13, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sundquist, K.; Palmér, K.; Hedelius, A.; Memon, A.A.; Sundquist, J. Macrophage Migration Inhibitory Factor and microRNA-451a in Response to Mindfulness-based Therapy or Treatment as Usual in Patients with Depression, Anxiety, or Stress and Adjustment Disorders. Int. J. Neuropsychopharmacol. 2018, 21, 513–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, S.; Li, L.; Song, L.; Wang, X. Effect of lamotrigine on cognitive function and serum inflammatory factors in patients with depression of recurrent bipolar disorder. Pak. J. Pharm. Sci. 2018, 31, 2775–2778. [Google Scholar] [PubMed]
- Benedetti, F.; Meazza, C.; Vivarelli, M.; Rossi, F.; Pistorio, A.; Lamb, R.; Lunt, M.; Thomson, W.; Ravelli, A.; Donn, R.; et al. Functional and prognostic relevance of the -173 polymorphism of the macrophage migration inhibitory factor gene in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2003, 48, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Baños-Hernández, C.J.; Navarro-Zarza, J.E.; Bucala, R.; Hernández-Bello, J.; Parra-Rojas, I.; Ramírez-Dueñas, M.G.; García-Arellano, S.; Hernández-Palma, L.A.; Machado-Sulbarán, A.C.; Muñoz-Valle, J.F. Macrophage migration inhibitory factor polymorphisms are a potential susceptibility marker in systemic sclerosis from southern Mexican population: Association with MIF mRNA expression and cytokine profile. Clin. Rheumatol. 2019, 38, 1643–1654. [Google Scholar] [CrossRef]
- Castañeda-Moreno, V.A.; La Cruz-Mosso, U.; Torres-Carrillo, N.; Macías-Islas, M.A.; La Padilla-De Torre, O.; Mireles-Ramírez, M.A.; González-Pérez, O.; Ruiz-Sandoval, J.L.; Huerta, M.; Trujillo, X.; et al. MIF functional polymorphisms (-794 CATT5-8 and -173 GC) are associated with MIF serum levels, severity and progression in male multiple sclerosis from western Mexican population. J. Neuroimmunol. 2018, 320, 117–124. [Google Scholar] [CrossRef]
- Radstake, T.R.D.J.; Sweep, F.C.G.J.; Welsing, P.; Franke, B.; Vermeulen, S.H.H.M.; Geurts-Moespot, A.; Calandra, T.; Donn, R.; van Riel, P.L.C.M. Correlation of rheumatoid arthritis severity with the genetic functional variants and circulating levels of macrophage migration inhibitory factor. Arthritis Rheum. 2005, 52, 3020–3029. [Google Scholar] [CrossRef]
- Yende, S.; Angus, D.C.; Kong, L.; Kellum, J.A.; Weissfeld, L.; Ferrell, R.; Finegold, D.; Carter, M.; Leng, L.; Peng, Z.-Y.; et al. The influence of macrophage migration inhibitory factor gene polymorphisms on outcome from community-acquired pneumonia. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2009, 23, 2403–2411. [Google Scholar] [CrossRef] [Green Version]
- Lao, W.; Xiang, Y.; Fang, M.; Yang, X. Polymorphism and expression of macrophage migration inhibitory factor does not contribute to glucocorticoid resistance in idiopathic thrombocytopenic purpura. Die Pharm. 2013, 68, 846–849. [Google Scholar]
- Matia-García, I.; Salgado-Goytia, L.; Muñoz-Valle, J.F.; García-Arellano, S.; Hernández-Bello, J.; Salgado-Bernabé, A.B.; Parra-Rojas, I. Macrophage migration inhibitory factor promoter polymorphisms (-794 CATT 5-8 and -173 GC): Relationship with mRNA expression and soluble MIF levels in young obese subjects. Dis. Mrk. 2015, 2015, 461208. [Google Scholar] [CrossRef]
- Jankauskas, S.S.; Wong, D.W.L.; Bucala, R.; Djudjaj, S.; Boor, P. Evolving complexity of MIF signaling. Cell. Signal. 2019, 57, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Florez-Sampedro, L.; Brandsma, C.A.; de Vries, M.; Timens, W.; Bults, R.; Vermeulen, C.J.; van den Berge, M.; Obeidat, M.; Joubert, P.; Nickle, D.C.; et al. Genetic regulation of gene expression of MIF family members in lung tissue. Sci. Rep. 2020, 10, 16980. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.W.; Bernhagen, J.; Bucala, R.; Lolis, E. Crystal structure at 2.6-A resolution of human macrophage migration inhibitory factor. Proc. Natl. Acad. Sci. USA 1996, 93, 5191–5196. [Google Scholar] [CrossRef] [Green Version]
- Rhein, C.; Tripal, P.; Seebahn, A.; Konrad, A.; Kramer, M.; Nagel, C.; Kemper, J.; Bode, J.; Muhle, C.; Gulbins, E.; et al. Functional implications of novel human acid sphingomyelinase splice variants. PLoS ONE 2012, 7, e35467. [Google Scholar] [CrossRef] [Green Version]
- Waeber, G.; Calandra, T.; Roduit, R.; Haefliger, J.A.; Bonny, C.; Thompson, N.; Thorens, B.; Temler, E.; Meinhardt, A.; Bacher, M.; et al. Insulin secretion is regulated by the glucose-dependent production of islet beta cell macrophage migration inhibitory factor. Proc. Natl. Acad. Sci. USA 1997, 94, 4782–4787. [Google Scholar] [CrossRef] [Green Version]
- Houdeau, E.; Moriez, R.; Leveque, M.; Salvador-Cartier, C.; Waget, A.; Leng, L.; Bueno, L.; Bucala, R.; Fioramonti, J. Sex steroid regulation of macrophage migration inhibitory factor in normal and inflamed colon in the female rat. Gastroenterology 2007, 132, 982–993. [Google Scholar] [CrossRef]
- von Zimmermann, C.; Bohm, L.; Richter-Schmidinger, T.; Kornhuber, J.; Lenz, B.; Muhle, C. Ex vivo glucocorticoid receptor-mediated IL-10 response predicts the course of depression severity. J. Neural Transm. 2021, 128, 95–104. [Google Scholar] [CrossRef]
- von Zimmermann, C.; Winkelmann, M.; Richter-Schmidinger, T.; Muhle, C.; Kornhuber, J.; Lenz, B. Physical Activity and Body Composition Are Associated With Severity and Risk of Depression, and Serum Lipids. Front. Psychiatry 2020, 11, 494. [Google Scholar] [CrossRef]
- Muhle, C.; Wagner, C.J.; Farber, K.; Richter-Schmidinger, T.; Gulbins, E.; Lenz, B.; Kornhuber, J. Secretory Acid Sphingomyelinase in the Serum of Medicated Patients Predicts the Prospective Course of Depression. J. Clin. Med. 2019, 8, 846. [Google Scholar] [CrossRef] [Green Version]
- Wagner, C.J.; Musenbichler, C.; Bohm, L.; Farber, K.; Fischer, A.I.; von Nippold, F.; Winkelmann, M.; Richter-Schmidinger, T.; Muhle, C.; Kornhuber, J.; et al. LDL cholesterol relates to depression, its severity, and the prospective course. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 92, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Sixth Revision of the Declaration of Helsinki. Available online: https://www.wma.net/wp-content/uploads/2018/07/DoH-Oct2008.pdf (accessed on 2 May 2013).
- International Conference on Harmonization Guidelines for Good Clinical Practice. Available online: https://www.ich.org/page/efficacy-guidelines (accessed on 2 May 2013).
- Hamilton, M. A rating scale for depression. J. Neurol. Neurosurg. Psychiatry 1960, 23, 56–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montgomery, S.A.; Asberg, M. A new depression scale designed to be sensitive to change. Br. J. Psychiatry J. Ment. Sci. 1979, 134, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.T.; Steer, R.A.; Brown, G.K. Manual for Beck Depression Inventory-II; Psychological Corporation: San Antonio, TX, USA, 1996. [Google Scholar]
- Lenz, B.; Klafki, H.-W.; Hillemacher, T.; Frieling, H.; Clepce, M.; Gossler, A.; Thuerauf, N.; Winterer, G.; Kornhuber, J.; Bleich, S. ERK1/2 protein and mRNA levels in human blood are linked to smoking behavior. Addict. Biol. 2012, 17, 1026–1035. [Google Scholar] [CrossRef]
- von Zimmermann, C.; Bruckner, L.; Muhle, C.; Weinland, C.; Kornhuber, J.; Lenz, B. Bioimpedance Body Measures and Serum Lipid Levels in Masculine Depression. Front. Psychiatry 2022, 13, 794351. [Google Scholar] [CrossRef]
| Unmedicated Patients | Premedicated Patients | Remitted Patients | Healthy Controls | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total Study Group | n | Median (IQR) | n | Median (IQR) | n | Median (IQR) | n | Median (IQR) | p Group | p Sex |
| Age (years) | 64 | 47 (34–53) | 66 | 46 (33–54) | 39 | 49 (46–58) | 61 | 42 (32–54) | 0.098 | 0.085 |
| Education (years) | 57 | 15 (13–18) | 58 | 14 (13–16) | 35 | 14 (13–16) | 51 | 15 (13–18) | 0.171 | 0.008 |
| BMI (kg/m2) | 64 | 25.2 (22.5–27.6) | 66 | 28.5 (24.4–30.4) | 39 | 25.7 (23.0–29.1) | 61 | 24.4 (23.0–27.7) | 0.001 | 0.003 |
| HAM-D T1 | 64 | 21 (19–24) | 66 | 23 (20–26) | 39 | 2 (0–3) | 61 | 1 (0–2) | <0.001 | 0.728 |
| HAM-D T2 | 60 | 18 (14–21) | 60 | 15 (10–22) | 0.189 | 0.072 | ||||
| HAM-D abs. change | 60 | −3.0 (−9.0–−1.0) | 60 | −7.5 (−11.0–−3.5) | 0.016 | 0.324 | ||||
| MADRS T1 | 64 | 26 (23–28) | 66 | 28 (24–34) | 39 | 1 (0–3) | 61 | 0 (0–2) | <0.001 | 0.890 |
| MADRS T2 | 60 | 21 (18–25) | 60 | 18 (13–26) | 0.143 | 0.072 | ||||
| MADRS abs. change | 60 | −4.5 (−8.5–−2.0) | 60 | −8.5 (−12.0–−4.0) | 0.009 | 0.038 | ||||
| BDI-II T1 | 64 | 28 (22–34) | 66 | 29 (24–35) | 39 | 3 (0–4) | 61 | 1 (0–3) | <0.001 | 0.609 |
| BDI-II T2 | 60 | 20 (15–25) | 60 | 20 (13–31) | 0.939 | 0.014 | ||||
| BDI-II abs. change | 60 | −8.5 (−11.0–−3.0) | 60 | −7.5 (−12.5–−2.0) | 0.975 | 0.220 | ||||
| MIF mRNA (AU) T1 | 63 | 0.07 (0.05–0.10) | 66 | 0.07 (0.05–0.10) | 38 | 0.07 (0.05–0.12) | 59 | 0.06 (0.03–0.10) | 0.316 | 0.003 |
| MIF mRNA (AU) T2 | 58 | 0.07 (0.06–0.12) | 60 | 0.06 (0.04–0.12) | 0.168 | 0.011 | ||||
| MIF mRNA rel. change | 57 | 0.06 (−0.32–0.66) | 60 | −0.15 (−0.40–0.61) | 0.502 | 0.471 | ||||
| MIF protein (pg/mL) T1. | 63 | 732 (602–1145) | 66 | 804 (586–1226) | 39 | 762 (529–1104) | 61 | 695 (567–919) | 0.590 | 0.003 |
| MIF protein (pg/mL) T2 | 60 | 699 (528–964) | 60 | 699 (571–1218) | 0.275 | 0.067 | ||||
| MIF protein rel. change | 60 | −0.07 (−0.25–0.10) | 60 | −0.06 (−0.19–0.14) | 0.513 | 0.741 | ||||
| Men | n | Median (IQR) | n | Median (IQR) | n | Median (IQR) | n | Median (IQR) | p Group | |
| Age (years) | 27 | 49 (35–53) | 34 | 46 (33–53) | 11 | 49 (33–53) | 30 | 37 (30–49) | 0.469 | |
| Education (years) | 23 | 16 (13–19) | 30 | 14 (13–16) | 10 | 15 (13–17) | 26 | 17 (14–18) | 0.127 | |
| BMI (kg/m2) | 27 | 25.7 (23.3–28.3) | 34 | 28.5 (26.7–30.2) | 11 | 25.8 (25.6–27.0) | 30 | 25.0 (22.9–28.4) | 0.005 | |
| HAM-D T1 | 27 | 21 (19–23) | 34 | 22 (20–25) | 11 | 2 (0–3) | 30 | 0 (0–1) | <0.001 | |
| HAM-D T2 | 26 | 18 (14–20) | 32 | 13 (9–21) | 0.173 | |||||
| HAM-D abs. change | 26 | −3.0 (−9.0–−1.0) | 32 | −9.0 (−11.0–−5.5) | 0.013 | |||||
| MADRS T1 | 27 | 27 (24–29) | 34 | 27 (23–34) | 11 | 2 (0–2) | 30 | 0 (0–1) | <0.001 | |
| MADRS T2 | 26 | 20 (18–24) | 32 | 17 (13–23) | 0.127 | |||||
| MADRS abs. change | 26 | −5.5 (−10.0–−2.0) | 32 | −9.5 (−13.0–−6.0) | 0.032 | |||||
| BDI-II T1 | 27 | 28 (23–32) | 34 | 27 (21–32) | 11 | 1 (0–3) | 30 | 2 (0–3) | <0.001 | |
| BDI-II T2 | 26 | 18 (15–22) | 32 | 17 (10–27) | 0.402 | |||||
| BDI-II abs. change | 26 | −9.0 (−12.0–−5.0) | 32 | −9.0 (−12.0–−3.5) | 0.987 | |||||
| MIF mRNA (AU) T1 | 26 | 0.06 (0.05–0.09) | 34 | 0.06 (0.05–0.09) | 10 | 0.09 (0.05–0.11) | 30 | 0.05 (0.03–0.08) | 0.224 | |
| MIF mRNA (AU) T2 | 25 | 0.06 (0.04–0.09) | 32 | 0.06 (0.04–0.11) | 0.664 | |||||
| MIF mRNA rel. change | 24 | 0.02 (−0.34–0.69) | 32 | −0.14 (−0.38–0.35) | 0.817 | |||||
| MIF protein (pg/mL) T1. | 27 | 900 (659–1454) | 34 | 783 (646–1120) | 11 | 859 (627–1425) | 30 | 746 (653–965) | 0.575 | |
| MIF protein (pg/mL) T2 | 27 | 775 (547–1254) | 32 | 704 (624–1165) | 0.976 | |||||
| MIF protein rel. change | 27 | −0.15 (−0.35–0.11) | 32 | −0.01 (−0.20–0.14) | 0.301 | |||||
| Women | n | Median (IQR) | n | Median (IQR) | n | Median (IQR) | n | Median (IQR) | p Group | |
| Age (years) | 37 | 45 (32–53) | 32 | 46 (32–56) | 28 | 52 (47–63) | 31 | 47 (32–60) | 0.082 | |
| Education (years) | 34 | 15 (13–17) | 28 | 14 (12–17) | 25 | 14 (12–15) | 25 | 14 (12–17) | 0.634 | |
| BMI (kg/m2) | 37 | 24.4 (21.7–27.3) | 32 | 27.3 (22.1–30.6) | 28 | 25.3 (22.7–29.2) | 31 | 24.3 (23.0–26.2) | 0.161 | |
| HAM-D T1 | 37 | 22 (19–25) | 32 | 24 (21–27) | 28 | 2 (1–4) | 31 | 1 (0–3) | <0.001 | |
| HAM-D T2 | 34 | 18 (14–21) | 28 | 17 (11–22) | 0.804 | |||||
| HAM-D abs. change | 34 | −4.5 (−9.0–−1.0) | 28 | −4.5 (−11.5–−2.0) | 0.474 | |||||
| MADRS T1 | 37 | 26 (22–28) | 32 | 28 (25–35) | 28 | 1 (0–4) | 31 | 1 (0–2) | <0.001 | |
| MADRS T2 | 34 | 21 (18–25) | 28 | 20 (15–29) | 0.755 | |||||
| MADRS abs. change | 34 | −4.0 (−8.0–−1.0) | 28 | −6.0 (−10.5–−2.0) | 0.201 | |||||
| BDI-II T1 | 37 | 29 (21–35) | 32 | 33 (27–39) | 28 | 3 (0–5) | 31 | 1 (0–4) | <0.001 | |
| BDI-II T2 | 34 | 22 (15–27) | 28 | 24 (16–36) | 0.318 | |||||
| BDI-II abs. change | 34 | −8.0 (−11.0–−1.0) | 28 | −6.0 (−13.0–0.0) | 0.921 | |||||
| MIF mRNA (AU) T1 | 37 | 0.08 (0.06–0.10) | 32 | 0.08 (0.05–0.14) | 28 | 0.07 (0.05–0.12) | 29 | 0.08 (0.06–0.10) | 0.956 | |
| MIF mRNA (AU) T2 | 33 | 0.08 (0.06–0.14) | 28 | 0.07 (0.05–0.16) | 0.347 | |||||
| MIF mRNA rel. change | 33 | 0.18 (−0.30–0.66) | 28 | −0.18 (−0.47–1.47) | 0.543 | |||||
| MIF protein (pg/mL) T1. | 36 | 664 (455–843) | 32 | 805 (509–1281) | 28 | 702 (492–1067) | 31 | 583 (480–807) | 0.364 | |
| MIF protein (pg/mL) T2 | 33 | 607 (447–823) | 28 | 662 (532–1290) | 0.230 | |||||
| MIF protein rel. change | 33 | −0.05 (−0.23–0.09) | 28 | −0.10 (−0.19–0.13) | 0.908 | |||||
| rs755622 | rs2070766 | rs2096525 | ||||||
|---|---|---|---|---|---|---|---|---|
| n | NN/Nn/nn | MAF | NN/Nn/nn | MAF | NN/Nn/nn | MAF | ||
| Remitted & current MDE patients (n = 168) | female | 96 | 63/33/0 | 0.172 | 66/30/0 | 0.156 | 65/31/0 | 0.161 |
| male | 72 | 46/24/2 | 0.194 | 48/23/1 | 0.174 | 46/23/3 | 0.201 | |
| Healthy control subjects (n = 61) | female | 31 | 19/8/4 | 0.258 | 20/8/3 | 0.226 | 20/6/5 | 0.258 |
| male | 30 | 18/11/1 | 0.217 | 19/10/1 | 0.200 | 20/9/1 | 0.183 | |
| Armitage’s trend test (NN + Nn vs. nn) | total | 229 | OR = 7.4 | p = 0.006 | OR = 11.7 | p = 0.006 | OR = 6.0 | p = 0.006 |
| female | 127 | OR = 31.6 | p < 0.001 | OR = 23.7 | p = 0.002 | OR = 40.1 | p < 0.001 | |
| male | 102 | OR = 1.2 | p = 0.880 | OR = 2.4 | p = 0.519 | OR = 0.8 | p = 0.843 |
| MIF Protein Level at Inclusion | Absolute Change of Score from Inclusion to Follow-Up | Sum Score at Follow-Up | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HAM-D | MADRS | BDI-II | HAM-D | MADRS | BDI-II | |||||||||
| n | rho | p | rho | p | rho | p | rho | p | rho | p | rho | p | ||
| Current MDE patients | All | 120 | −0.088 | 0.337 | −0.129 | 0.159 | 0.070 | 0.444 | −0.113 | 0.218 | −0.130 | 0.159 | −0.074 | 0.421 |
| Female | 62 | −0.106 | 0.414 | −0.111 | 0.391 | 0.123 | 0.341 | −0.067 | 0.605 | −0.084 | 0.516 | 0.026 | 0.842 | |
| Male | 58 | −0.006 | 0.966 | −0.066 | 0.622 | 0.041 | 0.760 | −0.096 | 0.473 | −0.127 | 0.343 | −0.084 | 0.533 | |
| Current MDE patients without premedication | All | 60 | 0.204 | 0.118 | 0.122 | 0.353 | 0.286 | 0.027 | 0.034 | 0.795 | 0.018 | 0.893 | 0.088 | 0.503 |
| Female | 34 | 0.194 | 0.272 | 0.226 | 0.199 | 0.382 | 0.026 | 0.030 | 0.866 | 0.100 | 0.575 | 0.231 | 0.188 | |
| Male | 26 | 0.135 | 0.510 | 0.101 | 0.622 | 0.099 | 0.330 | 0.090 | 0.663 | −0.069 | 0.737 | 0.029 | 0.889 | |
| Current MDE patients with premedication | All | 60 | −0.345 | 0.007 | −0.347 | 0.007 | −0.139 | 0.289 | −0.192 | 0.143 | −0.249 | 0.055 | −0.204 | 0.119 |
| Female | 28 | −0.354 | 0.065 | −0.426 | 0.024 | −0.118 | 0.550 | −0.185 | 0.345 | −0.287 | 0.138 | −0.265 | 0.174 | |
| Male | 32 | −0.236 | 0.194 | −0.304 | 0.090 | −0.175 | 0.338 | −0.198 | 0.278 | −0.245 | 0.176 | −0.154 | 0.401 | |
| MIF mRNA Level at Inclusion | Absolute Change of Score from Inclusion to Follow-Up | Sum Score at Follow-Up | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HAM-D | MADRS | BDI-II | HAM-D | MADRS | BDI-II | |||||||||
| n | rho | p | rho | p | rho | p | rho | p | rho | p | rho | p | ||
| Current MDE patients | All | 119 | 0.212 | 0.021 | 0.241 | 0.008 | 0.028 | 0.765 | 0.090 | 0.328 | 0.093 | 0.315 | 0.029 | 0.758 |
| Female | 62 | 0.211 | 0.100 | 0.234 | 0.067 | 0.003 | 0.979 | 0.192 | 0.135 | 0.191 | 0.137 | 0.065 | 0.617 | |
| Male | 57 | 0.201 | 0.134 | 0.178 | 0.186 | 0.027 | 0.841 | −0.049 | 0.758 | −0.056 | 0.676 | −0.072 | 0.595 | |
| Current MDE patients without premedication | All | 59 | 0.381 | 0.003 | 0.378 | 0.003 | 0.166 | 0.208 | 0.311 | 0.017 | 0.248 | 0.058 | 0.213 | 0.105 |
| Female | 34 | 0.428 | 0.012 | 0.439 | 0.003 | 0.171 | 0.335 | 0.383 | 0.025 | 0.284 | 0.104 | 0.173 | 0.328 | |
| Male | 25 | 0.309 | 0.132 | 0.176 | 0.401 | 0.130 | 0.536 | 0.168 | 0.423 | 0.166 | 0.428 | 0.279 | 0.177 | |
| Current MDE patients with premedication | All | 60 | −0.073 | 0.580 | 0.127 | 0.335 | −0.072 | 0.586 | −0.055 | 0.675 | −0.042 | 0.751 | −0.105 | 0.423 |
| Female | 28 | −0.057 | 0.772 | −0.060 | 0.760 | −0.213 | 0.277 | −0.011 | 0.956 | 0.041 | 0.835 | −0.069 | 0.728 | |
| Male | 32 | 0.162 | 0.377 | 0.183 | 0.315 | <0.001 | 0.998 | −0.146 | 0.426 | −0.150 | 0.413 | −0.204 | 0.262 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swoboda, C.; Deloch, L.; von Zimmermann, C.; Richter-Schmidinger, T.; Lenz, B.; Kornhuber, J.; Mühle, C. Macrophage Migration Inhibitory Factor in Major Depressive Disorder: A Multilevel Pilot Study. Int. J. Mol. Sci. 2022, 23, 15460. https://doi.org/10.3390/ijms232415460
Swoboda C, Deloch L, von Zimmermann C, Richter-Schmidinger T, Lenz B, Kornhuber J, Mühle C. Macrophage Migration Inhibitory Factor in Major Depressive Disorder: A Multilevel Pilot Study. International Journal of Molecular Sciences. 2022; 23(24):15460. https://doi.org/10.3390/ijms232415460
Chicago/Turabian StyleSwoboda, Caroline, Lena Deloch, Claudia von Zimmermann, Tanja Richter-Schmidinger, Bernd Lenz, Johannes Kornhuber, and Christiane Mühle. 2022. "Macrophage Migration Inhibitory Factor in Major Depressive Disorder: A Multilevel Pilot Study" International Journal of Molecular Sciences 23, no. 24: 15460. https://doi.org/10.3390/ijms232415460