Biochemical and Biophysical Characterization of the Caveolin-2 Interaction with Membranes and Analysis of the Protein Structural Alteration by the Presence of Cholesterol
Abstract
:1. Introduction
2. Results
2.1. Purification of Human Recombinant Cav-2
2.2. Structural Predictions Based on the Cav-2 Amino Acid Sequence
2.2.1. Homology Analysis
2.2.2. Prediction of Intrinsically Disordered Segments
2.2.3. Hydropathy Analysis
2.3. Far-UV-Circular Dichroism (CD) Spectra of Cav-2
2.4. Lipids Stabilize the Strucuture Cav-2
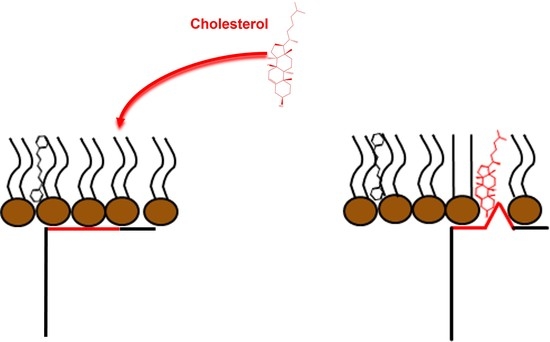
2.5. Cav-2 Interaction with Phospholipid Bilayers and Effect of Cholesterol
3. Discussion
4. Materials and Methods
4.1. Cav-2 Cloning
4.2. Cav-2 Expression in E. coli
4.3. Purification
4.3.1. Sephadex G75
4.3.2. Purification Using the His-Trap Affinity Column and Native Conditions
4.4. Preparation of SUVs and LUVs
4.5. Western Blotting
4.6. Circular Dichroism (CD)
4.7. Differential Scanning Calorimetry (DSC)
4.8. Fluorescence Intensity and Anisotropy Measurements
4.9. Prediction of Structural Information Obtained by Informatic Tools
4.10. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palade, G.E. Fine Structure of Blood Capillaries. J. Appl. Phys. 1953, 24, 1424. [Google Scholar]
- Stillwell, W. Chapter 11-Long-Range Membrane Properties. In An Introduction to Biological Membranes; Stillwell, W., Ed.; Elsevier: San Diego, CA, USA, 2013; pp. 215–237. ISBN 978-0-444-52153-8. [Google Scholar]
- Parton, R.G.; Hanzal-Bayer, M.; Hancock, J.F. Biogenesis of Caveolae: A Structural Model for Caveolin-Induced Domain Formation. J. Cell Sci. 2006, 119, 787–796. [Google Scholar] [CrossRef] [Green Version]
- Pol, A.; Morales-Paytuví, F.; Bosch, M.; Parton, R.G. Non-Caveolar Caveolins-Duties Outside the Caves. J. Cell Sci. 2020, 133, jcs241562. [Google Scholar] [CrossRef] [PubMed]
- Parton, R.G.; McMahon, K.-A.; Wu, Y. Caveolae: Formation, Dynamics, and Function. Curr. Opin. Cell Biol. 2020, 65, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Song, K.S.; Scherer, P.E.; Tang, Z.; Okamoto, T.; Li, S.; Chafel, M.; Chu, C.; Kohtz, D.S.; Lisanti, M.P. Expression of Caveolin-3 in Skeletal, Cardiac, and Smooth Muscle Cells. Caveolin-3 Is a Component of the Sarcolemma and Co-Fractionates with Dystrophin and Dystrophin-Associated Glycoproteins. J. Biol. Chem. 1996, 271, 15160–15165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sowa, G. Novel Insights into the Role of Caveolin-2 in Cell- and Tissue-Specific Signaling and Function. Biochem. Res. Int. 2011, 2011, 809259. [Google Scholar] [CrossRef] [Green Version]
- Williams, T.M.; Lisanti, M.P. The Caveolin Proteins. Genome Biol. 2004, 5, 214. [Google Scholar] [CrossRef] [Green Version]
- Scherer, P.E.; Okamoto, T.; Chun, M.; Nishimoto, I.; Lodish, H.F.; Lisanti, M.P. Identification, Sequence, and Expression of Caveolin-2 Defines a Caveolin Gene Family. Proc. Natl. Acad. Sci. USA 1996, 93, 131–135. [Google Scholar] [CrossRef] [Green Version]
- Collins, B.M.; Davis, M.J.; Hancock, J.F.; Parton, R.G. Structure-Based Reassessment of the Caveolin Signaling Model: Do Caveolae Regulate Signaling through Caveolin-Protein Interactions? Dev. Cell 2012, 23, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Byrne, D.P.; Dart, C.; Rigden, D.J. Evaluating Caveolin Interactions: Do Proteins Interact with the Caveolin Scaffolding Domain through a Widespread Aromatic Residue-Rich Motif? PLoS ONE 2012, 7, e44879. [Google Scholar] [CrossRef]
- Li, S.; Couet, J.; Lisanti, M.P. Src Tyrosine Kinases, Galpha Subunits, and H-Ras Share a Common Membrane-Anchored Scaffolding Protein, Caveolin. Caveolin Binding Negatively Regulates the Auto-Activation of Src Tyrosine Kinases. J. Biol. Chem. 1996, 271, 29182–29190. [Google Scholar] [CrossRef]
- Patel, H.H.; Murray, F.; Insel, P.A. Caveolae as Organizers of Pharmacologically Relevant Signal Transduction Molecules. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 359–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razani, B.; Woodman, S.E.; Lisanti, M.P. Caveolae: From Cell Biology to Animal Physiology. Pharmacol. Rev. 2002, 54, 431–467. [Google Scholar] [CrossRef]
- Yang, G.; Xu, H.; Li, Z.; Li, F. Interactions of Caveolin-1 Scaffolding and Intramembrane Regions Containing a CRAC Motif with Cholesterol in Lipid Bilayers. Biochim. Biophys. Acta 2014, 1838, 2588–2599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fantini, J.; Barrantes, F.J. How Cholesterol Interacts with Membrane Proteins: An Exploration of Cholesterol-Binding Sites Including CRAC, CARC, and Tilted Domains. Front. Physiol. 2013, 4, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baier, C.J.; Fantini, J.; Barrantes, F.J. Disclosure of Cholesterol Recognition Motifs in Transmembrane Domains of the Human Nicotinic Acetylcholine Receptor. Sci. Rep. 2011, 1, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, H.; Jeong, K.; Hwang, E.M.; Park, J.-Y.; Pak, Y. A Novel Domain of Caveolin-2 That Controls Nuclear Targeting: Regulation of Insulin-Specific ERK Activation and Nuclear Translocation by Caveolin-2. J. Cell. Mol. Med. 2011, 15, 888–908. [Google Scholar] [CrossRef] [Green Version]
- Breuza, L.; Corby, S.; Arsanto, J.-P.; Delgrossi, M.-H.; Scheiffele, P.; Le Bivic, A. The Scaffolding Domain of Caveolin 2 Is Responsible for Its Golgi Localization in Caco-2 Cells. J. Cell Sci. 2002, 115, 4457–4467. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, T.; Kogo, H.; Ishiguro, K.; Tauchi, K.; Nomura, R. Caveolin-2 Is Targeted to Lipid Droplets, a New “Membrane Domain” in the Cell. J. Cell Biol. 2001, 152, 1079–1085. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.; Hernandez, V.A.; Hu, K. Functional Interaction of the Two-Pore Domain Potassium Channel TASK-1 and Caveolin-3. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1537–1544. [Google Scholar] [CrossRef]
- Sotgia, F.; Lee, J.K.; Das, K.; Bedford, M.; Petrucci, T.C.; Macioce, P.; Sargiacomo, M.; Bricarelli, F.D.; Minetti, C.; Sudol, M.; et al. Caveolin-3 Directly Interacts with the C-Terminal Tail of Beta -Dystroglycan. Identification of a Central WW-like Domain within Caveolin Family Members. J. Biol. Chem. 2000, 275, 38048–38058. [Google Scholar] [CrossRef]
- Venema, V.J.; Ju, H.; Zou, R.; Venema, R.C. Interaction of Neuronal Nitric-Oxide Synthase with Caveolin-3 in Skeletal Muscle. Identification of a Novel Caveolin Scaffolding/Inhibitory Domain. J. Biol. Chem. 1997, 272, 28187–28190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volonte, D.; McTiernan, C.F.; Drab, M.; Kasper, M.; Galbiati, F. Caveolin-1 and Caveolin-3 Form Heterooligomeric Complexes in Atrial Cardiac Myocytes That Are Required for Doxorubicin-Induced Apoptosis. Am. J. Physiol. Heart Circ. Physiol. 2008, 294, H392–H401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Almeida, C.J.G. Caveolin-1 and Caveolin-2 Can Be Antagonistic Partners in Inflammation and Beyond. Front. Immunol. 2017, 8, 1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Okamoto, T.; Chun, M.; Sargiacomo, M.; Casanova, J.E.; Hansen, S.H.; Nishimoto, I.; Lisanti, M.P. Evidence for a Regulated Interaction between Heterotrimeric G Proteins and Caveolin. J. Biol. Chem. 1995, 270, 15693–15701. [Google Scholar] [CrossRef] [Green Version]
- Samhan-Arias, A.K.; Fortalezas, S.; Cordas, C.M.; Moura, I.; Moura, J.J.G.; Gutierrez-Merino, C. Cytochrome B5 Reductase Is the Component from Neuronal Synaptic Plasma Membrane Vesicles That Generates Superoxide Anion upon Stimulation by Cytochrome c. Redox Biol. 2018, 15, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Samhan-Arias, A.K.; Gutierrez-Merino, C. Purified NADH-Cytochrome B5 Reductase Is a Novel Superoxide Anion Source Inhibited by Apocynin: Sensitivity to Nitric Oxide and Peroxynitrite. Free Radic. Biol. Med. 2014, 73, 174–189. [Google Scholar] [CrossRef]
- Samhan-Arias, A.K.; Marques-da-Silva, D.; Yanamala, N.; Gutierrez-Merino, C. Stimulation and Clustering of Cytochrome B5 Reductase in Caveolin-Rich Lipid Microdomains Is an Early Event in Oxidative Stress-Mediated Apoptosis of Cerebellar Granule Neurons. J. Proteom. 2012, 75, 2934–2949. [Google Scholar] [CrossRef] [PubMed]
- Samhan-Arias, A.K.; Garcia-Bereguiain, M.A.; Martin-Romero, F.J.; Gutierrez-Merino, C. Clustering of Plasma Membrane-Bound Cytochrome B5 Reductase within “lipid Raft” Microdomains of the Neuronal Plasma Membrane. Mol. Cell. Neurosci. 2009, 40, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Valério, G.N.; Gutiérrez-Merino, C.; Nogueira, F.; Moura, I.; Moura, J.J.G.; Samhan-Arias, A.K. Human Erythrocytes Exposure to Juglone Leads to an Increase of Superoxide Anion Production Associated with Cytochrome B5 Reductase Uncoupling. Biochim. Biophys. Acta (BBA)-Bioenerg. 2020, 1861, 148134. [Google Scholar] [CrossRef]
- Mora, R.; Bonilha, V.L.; Marmorstein, A.; Scherer, P.E.; Brown, D.; Lisanti, M.P.; Rodriguez-Boulan, E. Caveolin-2 Localizes to the Golgi Complex but Redistributes to Plasma Membrane, Caveolae, and Rafts When Co-Expressed with Caveolin-1. J. Biol. Chem. 1999, 274, 25708–25717. [Google Scholar] [CrossRef]
- Ostermeyer, A.G.; Paci, J.M.; Zeng, Y.; Lublin, D.M.; Munro, S.; Brown, D.A. Accumulation of Caveolin in the Endoplasmic Reticulum Redirects the Protein to Lipid Storage Droplets. J. Cell Biol. 2001, 152, 1071–1078. [Google Scholar] [CrossRef] [Green Version]
- Burgermeister, E.; Tencer, L.; Liscovitch, M. Peroxisome Proliferator-Activated Receptor-Gamma Upregulates Caveolin-1 and Caveolin-2 Expression in Human Carcinoma Cells. Oncogene 2003, 22, 3888–3900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, H.; Jeong, K.; Pak, Y. Identification of PY19-Caveolin-2 as a Positive Regulator of Insulin-Stimulated Actin Cytoskeleton-Dependent Mitogenesis. J. Cell. Mol. Med. 2009, 13, 1549–1564. [Google Scholar] [CrossRef]
- Li, S.; Galbiati, F.; Volonte, D.; Sargiacomo, M.; Engelman, J.A.; Das, K.; Scherer, P.E.; Lisanti, M.P. Mutational Analysis of Caveolin-Induced Vesicle Formation. Expression of Caveolin-1 Recruits Caveolin-2 to Caveolae Membranes. FEBS Lett. 1998, 434, 127–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherer, P.E.; Lewis, R.Y.; Volonte, D.; Engelman, J.A.; Galbiati, F.; Couet, J.; Kohtz, D.S.; van Donselaar, E.; Peters, P.; Lisanti, M.P. Cell-Type and Tissue-Specific Expression of Caveolin-2. Caveolins 1 and 2 Co-Localize and Form a Stable Hetero-Oligomeric Complex in Vivo. J. Biol. Chem. 1997, 272, 29337–29346. [Google Scholar] [CrossRef] [Green Version]
- Song, K.S.; Tang, Z.; Li, S.; Lisanti, M.P. Mutational Analysis of the Properties of Caveolin-1. A Novel Role for the C-Terminal Domain in Mediating Homo-Typic Caveolin-Caveolin Interactions. J. Biol. Chem. 1997, 272, 4398–4403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sargiacomo, M.; Scherer, P.E.; Tang, Z.; Kübler, E.; Song, K.S.; Sanders, M.C.; Lisanti, M.P. Oligomeric Structure of Caveolin: Implications for Caveolae Membrane Organization. Proc. Natl. Acad. Sci. USA 1995, 92, 9407–9411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Lan, C.; Gallay, J.; Vincent, M.; Neumann, J.M.; de Foresta, B.; Jamin, N. Structural and Dynamic Properties of Juxta-Membrane Segments of Caveolin-1 and Caveolin-2 at the Membrane Interface. Eur. Biophys. J. 2010, 39, 307–325. [Google Scholar] [CrossRef]
- Mészáros, B.; Erdos, G.; Dosztányi, Z. IUPred2A: Context-Dependent Prediction of Protein Disorder as a Function of Redox State and Protein Binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef] [Green Version]
- Dosztányi, Z.; Mészáros, B.; Simon, I. ANCHOR: Web Server for Predicting Protein Binding Regions in Disordered Proteins. Bioinformatics 2009, 25, 2745–2746. [Google Scholar] [CrossRef]
- Miles, A.J.; Wallace, B.A. Circular Dichroism Spectroscopy of Membrane Proteins. Chem. Soc. Rev. 2016, 45, 4859–4872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjbar, B.; Gill, P. Circular Dichroism Techniques: Biomolecular and Nanostructural Analyses- a Review. Chem. Biol. Drug Des. 2009, 74, 101–120. [Google Scholar] [CrossRef] [PubMed]
- Andersen, N.H.; Liu, Z.; Prickett, K.S. Efforts toward Deriving the CD Spectrum of a 3(10) Helix in Aqueous Medium. FEBS Lett. 1996, 399, 47–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parton, D.L.; Klingelhoefer, J.W.; Sansom, M.S.P. Aggregation of Model Membrane Proteins, Modulated by Hydrophobic Mismatch, Membrane Curvature, and Protein Class. Biophys. J. 2011, 101, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stryer, L. Fluorescence Energy Transfer as a Spectroscopic Ruler. Annu. Rev. Biochem. 1978, 47, 819–846. [Google Scholar] [CrossRef]
- Aoki, S.; Thomas, A.; Decaffmeyer, M.; Brasseur, R.; Epand, R.M. The Role of Proline in the Membrane Re-Entrant Helix of Caveolin-1. J. Biol. Chem. 2010, 285, 33371–33380. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Glover, K.J. The Transmembrane Domain of Caveolin-1 Exhibits a Helix-Break-Helix Structure. Biochim. Biophys. Acta 2012, 1818, 1158–1164. [Google Scholar] [CrossRef] [Green Version]
- Spisni, E.; Tomasi, V.; Cestaro, A.; Tosatto, S.C.E. Structural Insights into the Function of Human Caveolin 1. Biochem. Biophys. Res. Commun. 2005, 338, 1383–1390. [Google Scholar] [CrossRef]
- Hoop, C.L.; Sivanandam, V.N.; Kodali, R.; Srnec, M.N.; van der Wel, P.C.A. Structural Characterization of the Caveolin Scaffolding Domain in Association with Cholesterol-Rich Membranes. Biochemistry 2012, 51, 90–99. [Google Scholar] [CrossRef]
- Schlegel, A.; Lisanti, M.P. A Molecular Dissection of Caveolin-1 Membrane Attachment and Oligomerization. Two Separate Regions of the Caveolin-1 C-Terminal Domain Mediate Membrane Binding and Oligomer/Oligomer Interactions in Vivo. J. Biol. Chem. 2000, 275, 21605–21617. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: New York, NY, USA, 2010; ISBN 978-0-387-31278-1. [Google Scholar]
- de Meyer, F.; Smit, B. Effect of Cholesterol on the Structure of a Phospholipid Bilayer. Proc. Natl. Acad. Sci. USA 2009, 106, 3654–3658. [Google Scholar] [CrossRef] [Green Version]
- Aguilar, L.F.; Pino, J.A.; Soto-Arriaza, M.A.; Cuevas, F.J.; Sánchez, S.; Sotomayor, C.P. Differential Dynamic and Structural Behavior of Lipid-Cholesterol Domains in Model Membranes. PLoS ONE 2012, 7, e40254. [Google Scholar] [CrossRef]
- Samhan-Arias, A.K.; Tyurina, Y.Y.; Kagan, V.E. Lipid Antioxidants: Free Radical Scavenging versus Regulation of Enzymatic Lipid Peroxidation. J. Clin. Biochem. Nutr. 2011, 48, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corpet, F. Multiple Sequence Alignment with Hierarchical Clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Snider, C.; Jayasinghe, S.; Hristova, K.; White, S.H. MPEx: A Tool for Exploring Membrane Proteins. Protein Sci. 2009, 18, 2624–2628. [Google Scholar] [CrossRef] [PubMed]






| STEP | Protein Amount | Yield (%) |
|---|---|---|
| Whole lysate Periplasmic fraction Sephadex G75 Niquel Sepharose | 530.1 | 100 |
| 182.0 | 34.3 | |
| 147.0 | 27.7 | |
| 4.2 | 1.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorospe, B.; Moura, J.J.G.; Gutierrez-Merino, C.; Samhan-Arias, A.K. Biochemical and Biophysical Characterization of the Caveolin-2 Interaction with Membranes and Analysis of the Protein Structural Alteration by the Presence of Cholesterol. Int. J. Mol. Sci. 2022, 23, 15203. https://doi.org/10.3390/ijms232315203
Gorospe B, Moura JJG, Gutierrez-Merino C, Samhan-Arias AK. Biochemical and Biophysical Characterization of the Caveolin-2 Interaction with Membranes and Analysis of the Protein Structural Alteration by the Presence of Cholesterol. International Journal of Molecular Sciences. 2022; 23(23):15203. https://doi.org/10.3390/ijms232315203
Chicago/Turabian StyleGorospe, Berta, José J. G. Moura, Carlos Gutierrez-Merino, and Alejandro K. Samhan-Arias. 2022. "Biochemical and Biophysical Characterization of the Caveolin-2 Interaction with Membranes and Analysis of the Protein Structural Alteration by the Presence of Cholesterol" International Journal of Molecular Sciences 23, no. 23: 15203. https://doi.org/10.3390/ijms232315203