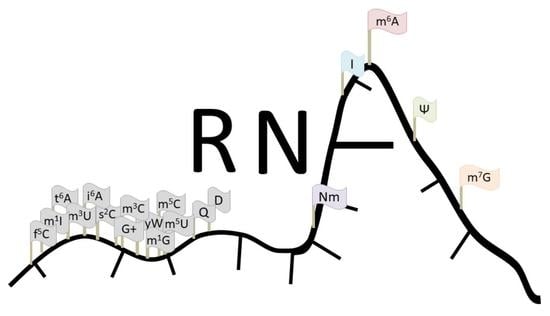
Epitranscriptome: Review of Top 25 Most-Studied RNA Modifications
Abstract
:1. Introduction
2. Current Status of Research on RNA Modifications
3. RNA Modifications at Large
4. RNA Modifications in Depth
4.1. N6-methyladenosine (m6A)
4.2. Inosine (I) and 1-methylinosine (m1I)
4.3. Pseudouridine (Ψ)
4.4. 5-methyluridine (m5U)
4.5. 2′-O-methylation of ribose (Nm or 2’-O-Me)
4.6. 1-methyladenosine (m1A)
4.7. Dihydrouridine
4.8. N4-acetylcytidine (ac4C)
4.9. Methyluridine (m3U)
4.10. 5-methylcytidine (m5C) and 5-formylcytidine (f5C)
4.11. 7-methylguanosine (m7G)
4.12. tRNA Structural Modifications
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Davis, F.F.; Allen, F.W. Ribonucleic Acids from Yeast Which Contain a Fifth Nucleotide. J. Biol. Chem. 1957, 227, 907–915. [Google Scholar] [CrossRef]
- Grosjean, H. RNA Modification: The Golden Period 1995–2015. RNA 2015, 21, 625–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frye, M.; Jaffrey, S.R.; Pan, T.; Rechavi, G.; Suzuki, T. RNA Modifications: What Have We Learned and Where Are We Headed? Nat. Rev. Genet. 2016, 17, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A Database of RNA Modification Pathways. 2021 Update. Nucleic Acids Res. 2022, 50, D231–D235. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.; Phan, L.; Cuesta, R.; Carlson, B.A.; Pak, M.; Asano, K.; Björk, G.R.; Tamame, M.; Hinnebusch, A.G. The Essential Gcd10p-Gcd14p Nuclear Complex Is Required for 1-Methyladenosine Modification and Maturation of Initiator Methionyl-TRNA. Genes Dev. 1998, 12, 3650–3662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, S.; Zhang, J.; Su, J.; Zuo, Z.; Zeng, L.; Liu, K.; Zheng, Y.; Huang, X.; Bai, R.; Zhuang, L.; et al. RNA M6A Regulates Transcription via DNA Demethylation and Chromatin Accessibility. Nat. Genet. 2022, 54, 1427–1437. [Google Scholar] [CrossRef]
- Zhou, K.I.; Shi, H.; Lyu, R.; Wylder, A.C.; Matuszek, Ż.; Pan, J.N.; He, C.; Parisien, M.; Pan, T. Regulation of Co-Transcriptional Pre-MRNA Splicing by M6A through the Low-Complexity Protein HnRNPG. Mol. Cell 2019, 76, 70–81.e9. [Google Scholar] [CrossRef]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA Modifications: Nature’s Combinatorial Chemistry Playground. Wiley Interdiscip. Rev. RNA 2013, 4, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Li, X.; Zhang, X.; Wang, Q.; Liu, W.; Lu, X.; Gao, S.; Liu, Z.; Liu, M.; Gao, L.; et al. RNA Motifs and Modification Involve in RNA Long-Distance Transport in Plants. Front. Cell Dev. Biol. 2021, 9, 651278. [Google Scholar] [CrossRef]
- Tong, J.; Zhang, W.; Chen, Y.; Yuan, Q.; Qin, N.-N.; Qu, G. The Emerging Role of RNA Modifications in the Regulation of Antiviral Innate Immunity. Front. Microbiol. 2022, 13, 845625. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m(6)A MRNA Methylation Directs Translational Control of Heat Shock Response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, C.T.Y.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A Quantitative Systems Approach Reveals Dynamic Control of TRNA Modifications during Cellular Stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef] [Green Version]
- Jonkhout, N.; Tran, J.; Smith, M.A.; Schonrock, N.; Mattick, J.S.; Novoa, E.M. The RNA Modification Landscape in Human Disease. RNA 2017, 23, 1754–1769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delaunay, S.; Frye, M. RNA Modifications Regulating Cell Fate in Cancer. Nat. Cell Biol. 2019, 21, 552–559. [Google Scholar] [CrossRef]
- Feng, Q.; Wang, D.; Xue, T.; Lin, C.; Gao, Y.; Sun, L.; Jin, Y.; Liu, D. The Role of RNA Modification in Hepatocellular Carcinoma. Front. Pharmacol. 2022, 13, 984453. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Puig, R.; Climent, F.; Piñeyro, D.; Domingo-Domènech, E.; Davalos, V.; Encuentra, M.; Rea, A.; Espejo-Herrera, N.; Soler, M.; Lopez, M.; et al. Epigenetic Loss of M1A RNA Demethylase ALKBH3 in Hodgkin Lymphoma Targets Collagen, Conferring Poor Clinical Outcome. Blood 2021, 137, 994–999. [Google Scholar] [CrossRef]
- Barbieri, I.; Kouzarides, T. Role of RNA Modifications in Cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef]
- Nance, K.D.; Meier, J.L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 748–756. [Google Scholar] [CrossRef]
- Ule, J.; Hwang, H.-W.; Darnell, R.B. The Future of Cross-Linking and Immunoprecipitation (CLIP). Cold Spring Harb. Perspect. Biol. 2018, 10, a032243. [Google Scholar] [CrossRef]
- Zou, S.; Toh, J.D.W.; Wong, K.H.Q.; Gao, Y.-G.; Hong, W.; Woon, E.C.Y. N(6)-Methyladenosine: A Conformational Marker That Regulates the Substrate Specificity of Human Demethylases FTO and ALKBH5. Sci. Rep. 2016, 6, 25677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oerum, S.; Meynier, V.; Catala, M.; Tisné, C. A Comprehensive Review of M6A/M6Am RNA Methyltransferase Structures. Nucleic Acids Res. 2021, 49, 7239–7255. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, D.A.; Sathe, S.; Einstein, J.M.; Yeo, G.W. Direct RNA Sequencing Enables M6A Detection in Endogenous Transcript Isoforms at Base-Specific Resolution. RNA 2020, 26, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Antanaviciute, A.; Baquero-Perez, B.; Watson, C.M.; Harrison, S.M.; Lascelles, C.; Crinnion, L.; Markham, A.F.; Bonthron, D.T.; Whitehouse, A.; Carr, I.M. M6aViewer: Software for the Detection, Analysis, and Visualization of N6-Methyladenosine Peaks from M6A-Seq/ME-RIP Sequencing Data. RNA 2017, 23, 1493–1501. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Hu, Y.; Zhou, B.; Bao, Y.; Li, Z.; Gong, C.; Yang, H.; Wang, S.; Xiao, Y. The Role of M6A Modification in Physiology and Disease. Cell Death Dis. 2020, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Li, H.-B.; Yin, Z.; Flavell, R.A. Recent Advances in Dynamic M6A RNA Modification. Open Biol. 2016, 6, 160003. [Google Scholar] [CrossRef] [Green Version]
- Torres, A.G.; Piñeyro, D.; Filonava, L.; Stracker, T.H.; Batlle, E.; Ribas de Pouplana, L. A-to-I Editing on TRNAs: Biochemical, Biological and Evolutionary Implications. FEBS Lett. 2014, 588, 4279–4286. [Google Scholar] [CrossRef]
- Srinivasan, S.; Torres, A.G.; Ribas de Pouplana, L. Inosine in Biology and Disease. Genes 2021, 12, 600. [Google Scholar] [CrossRef]
- Paul, M.S.; Bass, B.L. Inosine Exists in MRNA at Tissue-Specific Levels and Is Most Abundant in Brain MRNA. EMBO J. 1998, 17, 1120–1127. [Google Scholar] [CrossRef]
- Torres, A.G.; Piñeyro, D.; Rodríguez-Escribà, M.; Camacho, N.; Reina, O.; Saint-Léger, A.; Filonava, L.; Batlle, E.; Ribas de Pouplana, L. Inosine Modifications in Human TRNAs Are Incorporated at the Precursor TRNA Level. Nucleic Acids Res. 2015, 43, 5145–5157. [Google Scholar] [CrossRef] [Green Version]
- Torsin, L.I.; Petrescu, G.E.D.; Sabo, A.A.; Chen, B.; Brehar, F.M.; Dragomir, M.P.; Calin, G.A. Editing and Chemical Modifications on Non-Coding RNAs in Cancer: A New Tale with Clinical Significance. Int. J. Mol. Sci. 2021, 22, 581. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.A.; Heng, J.W.J.; Kaewsapsak, P.; Kok, E.P.L.; Stanojević, D.; Liu, H.; Cardilla, A.; Praditya, A.; Yi, Z.; Lin, M.; et al. Direct Identification of A-to-I Editing Sites with Nanopore Native RNA Sequencing. Nat. Methods 2022, 19, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Penzo, M.; Guerrieri, A.N.; Zacchini, F.; Treré, D.; Montanaro, L. RNA Pseudouridylation in Physiology and Medicine: For Better and for Worse. Genes 2017, 8, 301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteve-Puig, R.; Bueno-Costa, A.; Esteller, M. Writers, Readers and Erasers of RNA Modifications in Cancer. Cancer Lett. 2020, 474, 127–137. [Google Scholar] [CrossRef]
- Carlile, T.M.; Rojas-Duran, M.F.; Gilbert, W.V. Pseudo-Seq: Genome-Wide Detection of Pseudouridine Modifications in RNA. Methods Enzymol. 2015, 560, 219–245. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, A.F.; Riordan, D.P.; Brown, P.O. Transcriptome-Wide Mapping of Pseudouridines: Pseudouridine Synthases Modify Specific MRNAs in S. cerevisiae. PLoS ONE 2014, 9, e110799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhu, P.; Ma, S.; Song, J.; Bai, J.; Sun, F.; Yi, C. Chemical Pulldown Reveals Dynamic Pseudouridylation of the Mammalian Transcriptome. Nat. Chem. Biol. 2015, 11, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; León-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-Wide Mapping Reveals Widespread Dynamic-Regulated Pseudouridylation of NcRNA and MRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [Green Version]
- Khoddami, V.; Yerra, A.; Mosbruger, T.L.; Fleming, A.M.; Burrows, C.J.; Cairns, B.R. Transcriptome-Wide Profiling of Multiple RNA Modifications Simultaneously at Single-Base Resolution. Proc. Natl. Acad. Sci. USA 2019, 116, 6784–6789. [Google Scholar] [CrossRef]
- Garus, A.; Autexier, C. Dyskerin: An Essential Pseudouridine Synthase with Multifaceted Roles in Ribosome Biogenesis, Splicing, and Telomere Maintenance. RNA 2021, 27, 1441–1458. [Google Scholar] [CrossRef]
- Tomikawa, C. 7-Methylguanosine Modifications in Transfer RNA (TRNA). Int. J. Mol. Sci. 2018, 19, 4080. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Yao, Y.; Wu, P.; Zi, X.; Sun, N.; He, J. The Potential Role of N7-Methylguanosine (M7G) in Cancer. J. Hematol. Oncol. 2022, 15, 63. [Google Scholar] [CrossRef]
- Zhang, L.-S.; Liu, C.; Ma, H.; Dai, Q.; Sun, H.-L.; Luo, G.; Zhang, Z.; Zhang, L.; Hu, L.; Dong, X.; et al. Transcriptome-Wide Mapping of Internal N7-Methylguanosine Methylome in Mammalian MRNA. Mol. Cell 2019, 74, 1304–1316.e8. [Google Scholar] [CrossRef]
- Malbec, L.; Zhang, T.; Chen, Y.-S.; Zhang, Y.; Sun, B.-F.; Shi, B.-Y.; Zhao, Y.-L.; Yang, Y.; Yang, Y.-G. Dynamic Methylome of Internal MRNA N7-Methylguanosine and Its Regulatory Role in Translation. Cell Res. 2019, 29, 927–941. [Google Scholar] [CrossRef]
- Enroth, C.; Poulsen, L.D.; Iversen, S.; Kirpekar, F.; Albrechtsen, A.; Vinther, J. Detection of Internal N7-Methylguanosine (M7G) RNA Modifications by Mutational Profiling Sequencing. Nucleic Acids Res. 2019, 47, e126. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Kong, L.; Pei, Z.; Li, F.; Li, C.; Sun, X.; Shi, B.; Ge, J. M7G Methyltransferase METTL1 Promotes Post-Ischemic Angiogenesis via Promoting VEGFA MRNA Translation. Front. Cell Dev. Biol. 2021, 9, 642080. [Google Scholar] [CrossRef] [PubMed]
- Furlan, M.; Delgado-Tejedor, A.; Mulroney, L.; Pelizzola, M.; Novoa, E.M.; Leonardi, T. Computational Methods for RNA Modification Detection from Nanopore Direct RNA Sequencing Data. RNA Biol. 2021, 18, 31–40. [Google Scholar] [CrossRef]
- Kasprzak, J.M.; Czerwoniec, A.; Bujnicki, J.M. Molecular Evolution of Dihydrouridine Synthases. BMC Bioinform. 2012, 13, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Tanaka, Y.; Yamashita, K.; Suzuki, T.; Nakamura, A.; Hirano, N.; Suzuki, T.; Yao, M.; Tanaka, I. Molecular Basis of Dihydrouridine Formation on TRNA. Proc. Natl. Acad. Sci. USA 2011, 108, 19593–19598. [Google Scholar] [CrossRef] [PubMed]
- Draycott, A.S.; Schaening-Burgos, C.; Rojas-Duran, M.F.; Wilson, L.; Schärfen, L.; Neugebauer, K.M.; Nachtergaele, S.; Gilbert, W.V. Transcriptome-Wide Mapping Reveals a Diverse Dihydrouridine Landscape Including MRNA. PLoS Biol. 2022, 20, e3001622. [Google Scholar] [CrossRef]
- Finet, O.; Yague-Sanz, C.; Krüger, L.K.; Tran, P.; Migeot, V.; Louski, M.; Nevers, A.; Rougemaille, M.; Sun, J.; Ernst, F.G.M.; et al. Transcription-Wide Mapping of Dihydrouridine Reveals That MRNA Dihydrouridylation Is Required for Meiotic Chromosome Segregation. Mol. Cell 2022, 82, 404–419.e9. [Google Scholar] [CrossRef]
- Pomerantz, S.C.; McCloskey, J.A. Analysis of RNA Hydrolyzates by Liquid Chromatography-Mass Spectrometry. Methods Enzymol. 1990, 193, 796–824. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Li, X.; Xiong, X.; Wang, J.; Zhou, Z.; Zhu, X.; Gu, Y.; Dominissini, D.; He, L.; et al. N1-Methyladenosine Methylation in TRNA Drives Liver Tumourigenesis by Regulating Cholesterol Metabolism. Nat. Commun. 2021, 12, 6314. [Google Scholar] [CrossRef]
- Woo, H.-H.; Chambers, S.K. Human ALKBH3-Induced M1A Demethylation Increases the CSF-1 MRNA Stability in Breast and Ovarian Cancer Cells. Biochim. Biophys. Acta 2019, 1862, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Ueda, Y.; Ooshio, I.; Fusamae, Y.; Kitae, K.; Kawaguchi, M.; Jingushi, K.; Hase, H.; Harada, K.; Hirata, K.; Tsujikawa, K. AlkB Homolog 3-Mediated TRNA Demethylation Promotes Protein Synthesis in Cancer Cells. Sci. Rep. 2017, 7, 42271. [Google Scholar] [CrossRef] [Green Version]
- Cozen, A.E.; Quartley, E.; Holmes, A.D.; Hrabeta-Robinson, E.; Phizicky, E.M.; Lowe, T.M. ARM-Seq: AlkB-Facilitated RNA Methylation Sequencing Reveals a Complex Landscape of Modified TRNA Fragments. Nat. Methods 2015, 12, 879–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safra, M.; Sas-Chen, A.; Nir, R.; Winkler, R.; Nachshon, A.; Bar-Yaacov, D.; Erlacher, M.; Rossmanith, W.; Stern-Ginossar, N.; Schwartz, S. The M1A Landscape on Cytosolic and Mitochondrial MRNA at Single-Base Resolution. Nature 2017, 551, 251–255. [Google Scholar] [CrossRef]
- Dominissini, D.; Nachtergaele, S.; Moshitch-Moshkovitz, S.; Peer, E.; Kol, N.; Ben-Haim, M.S.; Dai, Q.; Di Segni, A.; Salmon-Divon, M.; Clark, W.C.; et al. The Dynamic N(1)-Methyladenosine Methylome in Eukaryotic Messenger RNA. Nature 2016, 530, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Hillmeier, M.; Wagner, M.; Ensfelder, T.; Korytiakova, E.; Thumbs, P.; Müller, M.; Carell, T. Synthesis and Structure Elucidation of the Human TRNA Nucleoside Mannosyl-Queuosine. Nat. Commun. 2021, 12, 7123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Pan, T. Quantitative Probing of Glycosylated Queuosine Modifications in TRNA. Methods Enzymol. 2021, 658, 73–82. [Google Scholar] [CrossRef]
- Pathak, C.; Jaiswal, Y.K.; Vinayak, M. Modulation in the Activity of Lactate Dehydrogenase and Level of C-Myc and c-Fos by Modified Base Queuine in Cancer. Cancer Biol. Ther. 2008, 7, 85–91. [Google Scholar] [CrossRef] [Green Version]
- Ishiwata, S.; Ozawa, Y.; Katayama, J.; Kaneko, S.; Shindo, H.; Tomioka, Y.; Ishiwata, T.; Asano, G.; Ikegawa, S.; Mizugaki, M. Elevated Expression Level of 60-KDa Subunit of TRNA-Guanine Transglycosylase in Colon Cancer. Cancer Lett. 2004, 212, 113–119. [Google Scholar] [CrossRef]
- Gu, X.; Liang, Z. Transcriptome-Wide Mapping 5-Methylcytosine by M5C RNA Immunoprecipitation Followed by Deep Sequencing in Plant. Methods Mol. Biol. 2019, 1933, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, A.; Sun, B.-F.; Yang, Y.; Han, Y.-N.; Yuan, X.; Chen, R.-X.; Wei, W.-S.; Liu, Y.; Gao, C.-C.; et al. 5-Methylcytosine Promotes Pathogenesis of Bladder Cancer through Stabilizing MRNAs. Nat. Cell Biol. 2019, 21, 978–990. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of MRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Liu, F.; Chen, W.; Miao, H.; Liang, H.; Liao, Z.; Zhang, Z.; Zhang, B. The Role of RNA M5C Modification in Cancer Metastasis. Int. J. Biol. Sci. 2021, 17, 3369–3380. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, C.; Spremulli, L.L.; Benkowski, L.A.; Ueda, T.; Yokogawa, T.; Watanabe, K. Unconventional Decoding of the AUA Codon as Methionine by Mitochondrial TRNAMet with the Anticodon F5CAU as Revealed with a Mitochondrial in Vitro Translation System. Nucleic Acids Res. 2009, 37, 1616–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriya, J.; Yokogawa, T.; Wakita, K.; Ueda, T.; Nishikawa, K.; Crain, P.F.; Hashizume, T.; Pomerantz, S.C.; McCloskey, J.A.; Kawai, G. A Novel Modified Nucleoside Found at the First Position of the Anticodon of Methionine TRNA from Bovine Liver Mitochondria. Biochemistry 1994, 33, 2234–2239. [Google Scholar] [CrossRef] [PubMed]
- Carter, J.-M.; Emmett, W.; Mozos, I.R.; Kotter, A.; Helm, M.; Ule, J.; Hussain, S. FICC-Seq: A Method for Enzyme-Specified Profiling of Methyl-5-Uridine in Cellular RNA. Nucleic Acids Res. 2019, 47, e113. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Song, B.; Tang, Y.; Chen, K.; Wei, Z.; Meng, J. M5UPred: A Web Server for the Prediction of RNA 5-Methyluridine Sites from Sequences. Mol. Ther. Nucleic Acids 2020, 22, 742–747. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Wang, H.; Shi, X.; Ye, L.; Yan, K.; Chen, Z.; Zhang, H.; Jin, Z.; Xue, X. Disease Activity-Associated Alteration of MRNA M5 C Methylation in CD4+ T Cells of Systemic Lupus Erythematosus. Front. Cell Dev. Biol. 2020, 8, 430. [Google Scholar] [CrossRef] [PubMed]
- Powell, C.A.; Minczuk, M. TRMT2B Is Responsible for Both TRNA and RRNA m 5 U-Methylation in Human Mitochondria. RNA Biol. 2020, 17, 451–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimitrova, D.G.; Teysset, L.; Carré, C. RNA 2′-O-Methylation (Nm) Modification in Human Diseases. Genes 2019, 10, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvanin, A.; Ayadi, L.; Helm, M.; Motorin, Y.; Marchand, V. Mapping and Quantification of TRNA 2′-O-Methylation by RiboMethSeq. Methods Mol. Biol. 2019, 1870, 273–295. [Google Scholar] [CrossRef]
- Incarnato, D.; Anselmi, F.; Morandi, E.; Neri, F.; Maldotti, M.; Rapelli, S.; Parlato, C.; Basile, G.; Oliviero, S. High-Throughput Single-Base Resolution Mapping of RNA 2′-O-Methylated Residues. Nucleic Acids Res. 2017, 45, 1433–1441. [Google Scholar] [CrossRef] [Green Version]
- Dai, Q.; Moshitch-Moshkovitz, S.; Han, D.; Kol, N.; Amariglio, N.; Rechavi, G.; Dominissini, D.; He, C. Nm-Seq Maps 2′-O-Methylation Sites in Human MRNA with Base Precision. Nat. Methods 2017, 14, 695–698. [Google Scholar] [CrossRef]
- Dong, Z.-W.; Shao, P.; Diao, L.-T.; Zhou, H.; Yu, C.-H.; Qu, L.-H. RTL-P: A Sensitive Approach for Detecting Sites of 2′-O-Methylation in RNA Molecules. Nucleic Acids Res. 2012, 40, e157. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Pirnie, S.P.; Carmichael, G.G. High-Throughput and Site-Specific Identification of 2′-O-Methylation Sites Using Ribose Oxidation Sequencing (RibOxi-Seq). RNA 2017, 23, 1303–1314. [Google Scholar] [CrossRef] [Green Version]
- Björk, G.R.; Jacobsson, K.; Nilsson, K.; Johansson, M.J.; Byström, A.S.; Persson, O.P. A Primordial TRNA Modification Required for the Evolution of Life? EMBO J. 2001, 20, 231–239. [Google Scholar] [CrossRef]
- Kempenaers, M.; Roovers, M.; Oudjama, Y.; Tkaczuk, K.L.; Bujnicki, J.M.; Droogmans, L. New Archaeal Methyltransferases Forming 1-Methyladenosine or 1-Methyladenosine and 1-Methylguanosine at Position 9 of TRNA. Nucleic Acids Res. 2010, 38, 6533–6543. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.; Franco, M.K.; Smith, T.J.; Snyder, L.R.; Anders, A.G.; Ruotolo, B.T.; Kennedy, R.T.; Koutmou, K.S. Methylated Guanosine and Uridine Modifications in S. cerevisiae MRNAs Modulate Translation Elongation. Biochemistry 2022. [Google Scholar] [CrossRef]
- Jin, X.; Lv, Z.; Gao, J.; Zhang, R.; Zheng, T.; Yin, P.; Li, D.; Peng, L.; Cao, X.; Qin, Y.; et al. AtTrm5a Catalyses 1-Methylguanosine and 1-Methylinosine Formation on TRNAs and Is Important for Vegetative and Reproductive Growth in Arabidopsis Thaliana. Nucleic Acids Res. 2019, 47, 883–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackman, J.E.; Montange, R.K.; Malik, H.S.; Phizicky, E.M. Identification of the Yeast Gene Encoding the TRNA M1G Methyltransferase Responsible for Modification at Position 9. RNA 2003, 9, 574–585. [Google Scholar] [CrossRef] [Green Version]
- Ranasinghe, R.T.; Challand, M.R.; Ganzinger, K.A.; Lewis, B.W.; Softley, C.; Schmied, W.H.; Horrocks, M.H.; Shivji, N.; Chin, J.W.; Spencer, J.; et al. Detecting RNA Base Methylations in Single Cells by in Situ Hybridization. Nat. Commun. 2018, 9, 655. [Google Scholar] [CrossRef] [Green Version]
- Leger, A.; Amaral, P.P.; Pandolfini, L.; Capitanchik, C.; Capraro, F.; Miano, V.; Migliori, V.; Toolan-Kerr, P.; Sideri, T.; Enright, A.J.; et al. RNA Modifications Detection by Comparative Nanopore Direct RNA Sequencing. Nat. Commun. 2021, 12, 7198. [Google Scholar] [CrossRef]
- Shippy, D.C.; Fadl, A.A. TRNA Modification Enzymes GidA and MnmE: Potential Role in Virulence of Bacterial Pathogens. Int. J. Mol. Sci. 2014, 15, 18267–18280. [Google Scholar] [CrossRef] [Green Version]
- Noma, A.; Sakaguchi, Y.; Suzuki, T. Mechanistic Characterization of the Sulfur-Relay System for Eukaryotic 2-Thiouridine Biogenesis at TRNA Wobble Positions. Nucleic Acids Res. 2009, 37, 1335–1352. [Google Scholar] [CrossRef] [Green Version]
- Vendeix, F.A.P.; Murphy, F.V.; Cantara, W.A.; Leszczyńska, G.; Gustilo, E.M.; Sproat, B.; Malkiewicz, A.; Agris, P.F. Human TRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism. J. Mol. Biol. 2012, 416, 467–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noma, A.; Kirino, Y.; Ikeuchi, Y.; Suzuki, T. Biosynthesis of Wybutosine, a Hyper-Modified Nucleoside in Eukaryotic Phenylalanine TRNA. EMBO J. 2006, 25, 2142–2154. [Google Scholar] [CrossRef] [PubMed]
- Perche-Letuvée, P.; Molle, T.; Forouhar, F.; Mulliez, E.; Atta, M. Wybutosine Biosynthesis: Structural and Mechanistic Overview. RNA Biol. 2014, 11, 1508–1518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, G.; Xu, M.; Zou, M.; Duan, S. The Processing, Gene Regulation, Biological Functions, and Clinical Relevance of N4-Acetylcytidine on RNA: A Systematic Review. Mol. Ther. Nucleic Acids 2020, 20, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Sas-Chen, A.; Thomas, J.M.; Matzov, D.; Taoka, M.; Nance, K.D.; Nir, R.; Bryson, K.M.; Shachar, R.; Liman, G.L.S.; Burkhart, B.W.; et al. Dynamic RNA Acetylation Revealed by Quantitative Cross-Evolutionary Mapping. Nature 2020, 583, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Arango, D.; Sturgill, D.; Alhusaini, N.; Dillman, A.A.; Sweet, T.J.; Hanson, G.; Hosogane, M.; Sinclair, W.R.; Nanan, K.K.; Mandler, M.D.; et al. Acetylation of Cytidine in MRNA Promotes Translation Efficiency. Cell 2018, 175, 1872–1886.e24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tardu, M.; Jones, J.D.; Kennedy, R.T.; Lin, Q.; Koutmou, K.S. Identification and Quantification of Modified Nucleosides in Saccharomyces Cerevisiae MRNAs. ACS Chem. Biol. 2019, 14, 1403–1409. [Google Scholar] [CrossRef]
- Bohnsack, K.E.; Kleiber, N.; Lemus-Diaz, N.; Bohnsack, M.T. Roles and Dynamics of 3-Methylcytidine in Cellular RNAs. Trends Biochem. Sci. 2022, 47, 596–608. [Google Scholar] [CrossRef]
- Marchand, V.; Ayadi, L.; Bourguignon-Igel, V.; Helm, M.; Motorin, Y. AlkAniline-Seq: A Highly Sensitive and Specific Method for Simultaneous Mapping of 7-Methyl-Guanosine (M7G) and 3-Methyl-Cytosine (M3C) in RNAs by High-Throughput Sequencing. Methods Mol. Biol. 2021, 2298, 77–95. [Google Scholar] [CrossRef]
- Cui, J.; Liu, Q.; Sendinc, E.; Shi, Y.; Gregory, R.I. Nucleotide Resolution Profiling of M3C RNA Modification by HAC-Seq. Nucleic Acids Res. 2021, 49, e27. [Google Scholar] [CrossRef]
- Zheng, G.; Qin, Y.; Clark, W.C.; Dai, Q.; Yi, C.; He, C.; Lambowitz, A.M.; Pan, T. Efficient and Quantitative High-Throughput TRNA Sequencing. Nat. Methods 2015, 12, 835–837. [Google Scholar] [CrossRef] [PubMed]
- Behrens, A.; Rodschinka, G.; Nedialkova, D.D. High-Resolution Quantitative Profiling of TRNA Abundance and Modification Status in Eukaryotes by Mim-TRNAseq. Mol. Cell 2021, 81, 1802–1815.e7. [Google Scholar] [CrossRef] [PubMed]
- Gogakos, T.; Brown, M.; Garzia, A.; Meyer, C.; Hafner, M.; Tuschl, T. Characterizing Expression and Processing of Precursor and Mature Human TRNAs by Hydro-TRNAseq and PAR-CLIP. Cell Rep. 2017, 20, 1463–1475. [Google Scholar] [CrossRef] [Green Version]
- Yokogawa, T.; Nomura, Y.; Yasuda, A.; Ogino, H.; Hiura, K.; Nakada, S.; Oka, N.; Ando, K.; Kawamura, T.; Hirata, A.; et al. Identification of a Radical SAM Enzyme Involved in the Synthesis of Archaeosine. Nat. Chem. Biol. 2019, 15, 1148–1155. [Google Scholar] [CrossRef]
- Turner, B.; Burkhart, B.W.; Weidenbach, K.; Ross, R.; Limbach, P.A.; Schmitz, R.A.; de Crécy-Lagard, V.; Stedman, K.M.; Santangelo, T.J.; Iwata-Reuyl, D. Archaeosine Modification of Archaeal TRNA: Role in Structural Stabilization. J. Bacteriol. 2020, 202, e00748-19. [Google Scholar] [CrossRef] [PubMed]
- Lentini, J.M.; Fu, D. Monitoring the 5-Methoxycarbonylmethyl-2-Thiouridine (Mcm5s2U) Modification Utilizing the Gamma-Toxin Endonuclease. Methods Mol. Biol. 2021, 2298, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Jäger, G.; Leipuviene, R.; Pollard, M.G.; Qian, Q.; Björk, G.R. The Conserved Cys-X1-X2-Cys Motif Present in the TtcA Protein Is Required for the Thiolation of Cytidine in Position 32 of TRNA from Salmonella Enterica Serovar Typhimurium. J. Bacteriol. 2004, 186, 750–757. [Google Scholar] [CrossRef] [Green Version]
- Vangaveti, S.; Cantara, W.A.; Spears, J.L.; DeMirci, H.; Murphy, F.V.; Ranganathan, S.V.; Sarachan, K.L.; Agris, P.F. A Structural Basis for Restricted Codon Recognition Mediated by 2-Thiocytidine in TRNA Containing a Wobble Position Inosine. J. Mol. Biol. 2020, 432, 913–929. [Google Scholar] [CrossRef]
- Hirata, A.; Suzuki, T.; Nagano, T.; Fujii, D.; Okamoto, M.; Sora, M.; Lowe, T.M.; Kanai, T.; Atomi, H.; Suzuki, T.; et al. Distinct Modified Nucleosides in TRNATrp from the Hyperthermophilic Archaeon Thermococcus Kodakarensis and Requirement of TRNA M2G10/M2 2G10 Methyltransferase (Archaeal Trm11) for Survival at High Temperatures. J. Bacteriol. 2019, 201, e00448-19. [Google Scholar] [CrossRef]
- Bavi, R.S.; Sambhare, S.B.; Sonawane, K.D. MD Simulation Studies to Investigate Iso-Energetic Conformational Behaviour of Modified Nucleosides m(2)G and m(2) 2G Present in TRNA. Comput. Struct. Biotechnol. J. 2013, 5, e201302015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamichhane, T.N.; Arimbasseri, A.G.; Rijal, K.; Iben, J.R.; Wei, F.Y.; Tomizawa, K.; Maraia, R.J. Lack of TRNA-I6A Modification Causes Mitochondrial-like Metabolic Deficiency in S. Pombe by Limiting Activity of Cytosolic TRNATyr, Not Mito-TRNA. RNA 2016, 22, 583–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schweizer, U.; Bohleber, S.; Fradejas-Villar, N. The Modified Base Isopentenyladenosine and Its Derivatives in TRNA. RNA Biol. 2017, 14, 1197–1208. [Google Scholar] [CrossRef]
- Pereira, M.; Francisco, S.; Varanda, A.S.; Santos, M.; Santos, M.A.S.; Soares, A.R. Impact of TRNA Modifications and TRNA-Modifying Enzymes on Proteostasis and Human Disease. Int. J. Mol. Sci. 2018, 19, 3738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrochia, L.; Crozat, E.; Hecker, A.; Zhang, W.; Bareille, J.; Collinet, B.; van Tilbeurgh, H.; Forterre, P.; Basta, T. In Vitro Biosynthesis of a Universal T6A TRNA Modification in Archaea and Eukarya. Nucleic Acids Res. 2013, 41, 1953–1964. [Google Scholar] [CrossRef] [Green Version]
- Berulava, T.; Ziehe, M.; Klein-Hitpass, L.; Mladenov, E.; Thomale, J.; Rüther, U.; Horsthemke, B. FTO Levels Affect RNA Modification and the Transcriptome. Eur. J. Hum. Genet. 2013, 21, 317–323. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Yang, J.; Düttmann, S.; Watzinger, P.; Kötter, P.; Entian, K.-D. Identification of Novel Methyltransferases, Bmt5 and Bmt6, Responsible for the M3U Methylations of 25S RRNA in Saccharomyces Cerevisiae. Nucleic Acids Res. 2014, 42, 3246–3260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Crécy-Lagard, V.; Brochier-Armanet, C.; Urbonavicius, J.; Fernandez, B.; Phillips, G.; Lyons, B.; Noma, A.; Alvarez, S.; Droogmans, L.; Armengaud, J.; et al. Biosynthesis of Wyosine Derivatives in TRNA: An Ancient and Highly Diverse Pathway in Archaea. Mol. Biol. Evol. 2010, 27, 2062–2077. [Google Scholar] [CrossRef] [Green Version]
- Urbonavičius, J.; Tauraitė, D. Biochemical Pathways Leading to the Formation of Wyosine Derivatives in TRNA of Archaea. Biomolecules 2020, 10, 1627. [Google Scholar] [CrossRef]
- Dutta, N.; Deb, I.; Sarzynska, J.; Lahiri, A. Inosine and Its Methyl Derivatives: Occurrence, Biogenesis, and Function in RNA. Prog. Biophys. Mol. Biol. 2022, 169–170, 21–52. [Google Scholar] [CrossRef]
- Tanzer, A.; Hofacker, I.L.; Lorenz, R. RNA Modifications in Structure Prediction—Status Quo and Future Challenges. Methods 2019, 156, 32–39. [Google Scholar] [CrossRef]
- Wolk, S.K.; Mayfield, W.S.; Gelinas, A.D.; Astling, D.; Guillot, J.; Brody, E.N.; Janjic, N.; Gold, L. Modified Nucleotides May Have Enhanced Early RNA Catalysis. Proc. Natl. Acad. Sci. USA 2020, 117, 8236–8242. [Google Scholar] [CrossRef]
- Taoka, M.; Nobe, Y.; Yamaki, Y.; Sato, K.; Ishikawa, H.; Izumikawa, K.; Yamauchi, Y.; Hirota, K.; Nakayama, H.; Takahashi, N.; et al. Landscape of the Complete RNA Chemical Modifications in the Human 80S Ribosome. Nucleic Acids Res. 2018, 46, 9289–9298. [Google Scholar] [CrossRef] [PubMed]
- Legrand, C.; Tuorto, F.; Hartmann, M.; Liebers, R.; Jacob, D.; Helm, M.; Lyko, F. Statistically Robust Methylation Calling for Whole-Transcriptome Bisulfite Sequencing Reveals Distinct Methylation Patterns for Mouse RNAs. Genome Res. 2017, 27, 1589–1596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoernes, T.P.; Hüttenhofer, A.; Erlacher, M.D. MRNA Modifications: Dynamic Regulators of Gene Expression? RNA Biol. 2016, 13, 760–765. [Google Scholar] [CrossRef] [Green Version]
- Tan, K.-T.; Ding, L.-W.; Wu, C.-S.; Tenen, D.G.; Yang, H. Repurposing RNA Sequencing for Discovery of RNA Modifications in Clinical Cohorts. Sci. Adv. 2021, 7, eabd2605. [Google Scholar] [CrossRef]
- Shelton, S.B.; Reinsborough, C.; Xhemalce, B. Who Watches the Watchmen: Roles of RNA Modifications in the RNA Interference Pathway. PLoS Genet. 2016, 12, e1006139. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-H.; Wang, R.; Xiong, F.; Krakowiak, J.; Liao, Z.; Nguyen, P.T.; Moroz-Omori, E.V.; Shao, J.; Zhu, X.; Bolt, M.J.; et al. Enhancer RNA M6A Methylation Facilitates Transcriptional Condensate Formation and Gene Activation. Mol. Cell 2021, 81, 3368–3385.e9. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Ding, Q.; Xie, D.; Cai, Z.; Zhao, Z. Technical Challenges in Defining RNA Modifications. Semin. Cell Dev. Biol. 2022, 127, 155–165. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic Transcriptomic M6A Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Y.; Dong, L.; Liu, X.-M.; Guo, J.; Ma, H.; Shen, B.; Qian, S.-B. M6A in MRNA Coding Regions Promotes Translation via the RNA Helicase-Containing YTHDC2. Nat. Commun. 2019, 10, 5332. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Hao, Y.-J.; Zhang, Y.; Li, M.-M.; Wang, M.; Han, W.; Wu, Y.; Lv, Y.; Hao, J.; Wang, L.; et al. M(6)A RNA Methylation Is Regulated by MicroRNAs and Promotes Reprogramming to Pluripotency. Cell Stem Cell 2015, 16, 289–301. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The Role of M6A Modification in the Biological Functions and Diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Ren, W.; Lu, J.; Huang, M.; Gao, L.; Li, D.; Wang, G.G.; Song, J. Structure and Regulation of ZCCHC4 in M6A-Methylation of 28S RRNA. Nat. Commun. 2019, 10, 5042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 2015, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blazie, S.M.; Takayanagi-Kiya, S.; McCulloch, K.A.; Jin, Y. Eukaryotic Initiation Factor EIF-3.G Augments MRNA Translation Efficiency to Regulate Neuronal Activity. eLife 2021, 10, e68336. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the Human and Mouse M6A RNA Methylomes Revealed by M6A-Seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Grozhik, A.V.; Linder, B.; Olarerin-George, A.O.; Jaffrey, S.R. Mapping M6A at Individual-Nucleotide Resolution Using Crosslinking and Immunoprecipitation (MiCLIP). Methods Mol. Biol. 2017, 1562, 55–78. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Lu, Z.; Wang, X.; Fu, Y.; Luo, G.-Z.; Liu, N.; Han, D.; Dominissini, D.; Dai, Q.; Pan, T.; et al. High-Resolution N(6) -Methyladenosine (m(6) A) Map Using Photo-Crosslinking-Assisted m(6) A Sequencing. Angew. Chem. Int. Ed. 2015, 54, 1587–1590. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Xiong, X.; Yi, C. Epitranscriptome Sequencing Technologies: Decoding RNA Modifications. Nat. Methods 2016, 14, 23–31. [Google Scholar] [CrossRef]
- Deng, L.-J.; Deng, W.-Q.; Fan, S.-R.; Chen, M.-F.; Qi, M.; Lyu, W.-Y.; Qi, Q.; Tiwari, A.K.; Chen, J.-X.; Zhang, D.-M.; et al. M6A Modification: Recent Advances, Anticancer Targeted Drug Discovery and Beyond. Mol. Cancer 2022, 21, 52. [Google Scholar] [CrossRef]
- Gott, J.M.; Emeson, R.B. Functions and Mechanisms of RNA Editing. Annu. Rev. Genet. 2000, 34, 499–531. [Google Scholar] [CrossRef]
- Bazak, L.; Haviv, A.; Barak, M.; Jacob-Hirsch, J.; Deng, P.; Zhang, R.; Isaacs, F.J.; Rechavi, G.; Li, J.B.; Eisenberg, E.; et al. A-to-I RNA Editing Occurs at over a Hundred Million Genomic Sites, Located in a Majority of Human Genes. Genome Res. 2014, 24, 365–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rafels-Ybern, À.; Torres, A.G.; Grau-Bove, X.; Ruiz-Trillo, I.; Ribas de Pouplana, L. Codon Adaptation to TRNAs with Inosine Modification at Position 34 Is Widespread among Eukaryotes and Present in Two Bacterial Phyla. RNA Biol. 2018, 15, 500–507. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Karcher, D.; Bock, R. Importance of Adenosine-to-Inosine Editing Adjacent to the Anticodon in an Arabidopsis Alanine TRNA under Environmental Stress. Nucleic Acids Res. 2013, 41, 3362–3372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edmonds, C.G.; Crain, P.F.; Gupta, R.; Hashizume, T.; Hocart, C.H.; Kowalak, J.A.; Pomerantz, S.C.; Stetter, K.O.; McCloskey, J.A. Posttranscriptional Modification of TRNA in Thermophilic Archaea (Archaebacteria). J. Bacteriol. 1991, 173, 3138–3148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishikura, K. A-to-I Editing of Coding and Non-Coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Zhang, W.; Li, Q. Origins and Evolution of ADAR-Mediated RNA Editing. IUBMB Life 2009, 61, 572–578. [Google Scholar] [CrossRef]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic Identification of Abundant A-to-I Editing Sites in the Human Transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005. [Google Scholar] [CrossRef] [Green Version]
- Hundley, H.A.; Bass, B.L. ADAR Editing in Double-Stranded UTRs and Other Noncoding RNA Sequences. Trends Biochem. Sci. 2010, 35, 377–383. [Google Scholar] [CrossRef] [Green Version]
- Wright, A.; Vissel, B. The Essential Role of AMPA Receptor GluR2 Subunit RNA Editing in the Normal and Diseased Brain. Front. Mol. Neurosci. 2012, 5, 34. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Okada, S.; Sakurai, M. Adenosine-to-Inosine RNA Editing in Neurological Development and Disease. RNA Biol. 2021, 18, 999–1013. [Google Scholar] [CrossRef]
- Levitsky, L.I.; Kliuchnikova, A.A.; Kuznetsova, K.G.; Karpov, D.S.; Ivanov, M.V.; Pyatnitskiy, M.A.; Kalinina, O.V.; Gorshkov, M.V.; Moshkovskii, S.A. Adenosine-to-Inosine RNA Editing in Mouse and Human Brain Proteomes. Proteomics 2019, 19, e1900195. [Google Scholar] [CrossRef] [PubMed]
- Kliuchnikova, A.A.; Goncharov, A.O.; Levitsky, L.I.; Pyatnitskiy, M.A.; Novikova, S.E.; Kuznetsova, K.G.; Ivanov, M.V.; Ilina, I.Y.; Farafonova, T.E.; Zgoda, V.G.; et al. Proteome-Wide Analysis of ADAR-Mediated Messenger RNA Editing during Fruit Fly Ontogeny. J. Proteome Res. 2020, 19, 4046–4060. [Google Scholar] [CrossRef] [PubMed]
- Katrekar, D.; Chen, G.; Meluzzi, D.; Ganesh, A.; Worlikar, A.; Shih, Y.-R.; Varghese, S.; Mali, P. In Vivo RNA Editing of Point Mutations via RNA-Guided Adenosine Deaminases. Nat. Methods 2019, 16, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Reardon, S. Step aside CRISPR, RNA Editing Is Taking Off. Nature 2020, 578, 24–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohn, W.E.; Volkin, E. Nucleoside-5′-Phosphates from Ribonucleic Acid. Nature 1951, 167, 483–484. [Google Scholar] [CrossRef]
- Rintala-Dempsey, A.C.; Kothe, U. Eukaryotic Stand-Alone Pseudouridine Synthases—RNA Modifying Enzymes and Emerging Regulators of Gene Expression? RNA Biol. 2017, 14, 1185–1196. [Google Scholar] [CrossRef] [Green Version]
- Roovers, M.; Droogmans, L.; Grosjean, H. Post-Transcriptional Modifications of Conserved Nucleotides in the T-Loop of TRNA: A Tale of Functional Convergent Evolution. Genes 2021, 12, 140. [Google Scholar] [CrossRef]
- Roovers, M.; Hale, C.; Tricot, C.; Terns, M.P.; Terns, R.M.; Grosjean, H.; Droogmans, L. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res. 2006, 34, 4293–4301. [Google Scholar] [CrossRef]
- Becker, H.F.; Motorin, Y.; Planta, R.J.; Grosjean, H. The Yeast Gene YNL292w Encodes a Pseudouridine Synthase (Pus4) Catalyzing the Formation of Psi55 in Both Mitochondrial and Cytoplasmic TRNAs. Nucleic Acids Res. 1997, 25, 4493–4499. [Google Scholar] [CrossRef]
- Davis, D.R. Stabilization of RNA Stacking by Pseudouridine. Nucleic Acids Res. 1995, 23, 5020–5026. [Google Scholar] [CrossRef]
- Liang, X.-H.; Liu, Q.; Fournier, M.J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009, 15, 1716–1728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzzi, N.; Muthukumar, S.; Cieśla, M.; Todisco, G.; Ngoc, P.C.T.; Madej, M.; Munita, R.; Fazio, S.; Ekström, S.; Mortera-Blanco, T.; et al. Pseudouridine-Modified TRNA Fragments Repress Aberrant Protein Synthesis and Predict Leukaemic Progression in Myelodysplastic Syndrome. Nat. Cell Biol. 2022, 24, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Bykhovskaya, Y.; Casas, K.; Mengesha, E.; Inbal, A.; Fischel-Ghodsian, N. Missense Mutation in Pseudouridine Synthase 1 (PUS1) Causes Mitochondrial Myopathy and Sideroblastic Anemia (MLASA). Am. J. Hum. Genet. 2004, 74, 1303–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubin, D.T. Methylated Nucleotide Content of Mitochondrial Ribosomal RNA from Hamster Cells. J. Mol. Biol. 1974, 84, 257–273. [Google Scholar] [CrossRef]
- Desmolaize, B.; Fabret, C.; Brégeon, D.; Rose, S.; Grosjean, H.; Douthwaite, S. A Single Methyltransferase YefA (RlmCD) Catalyses Both M5U747 and M5U1939 Modifications in Bacillus Subtilis 23S RRNA. Nucleic Acids Res. 2011, 39, 9368–9375. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.-H.; Nishimura, S.; Oishi, H.; Kelly, V.P.; Kuno, A.; Takahashi, S. TRMT2A Is a Novel Cell Cycle Regulator That Suppresses Cell Proliferation. Biochem. Biophys. Res. Commun. 2019, 508, 410–415. [Google Scholar] [CrossRef]
- Ny, T.; Björk, G.R. Cloning and Restriction Mapping of the TrmA Gene Coding for Transfer Ribonucleic Acid (5-Methyluridine)-Methyltransferase in Escherichia Coli K-12. J. Bacteriol. 1980, 142, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Nordlund, M.E.; Johansson, J.O.; von Pawel-Rammingen, U.; Byström, A.S. Identification of the TRM2 Gene Encoding the TRNA(M5U54)Methyltransferase of Saccharomyces Cerevisiae. RNA 2000, 6, 844–860. [Google Scholar] [CrossRef] [Green Version]
- Pereira, M.; Ribeiro, D.R.; Pinheiro, M.M.; Ferreira, M.; Kellner, S.; Soares, A.R. M5U54 TRNA Hypomodification by Lack of TRMT2A Drives the Generation of TRNA-Derived Small RNAs. Int. J. Mol. Sci. 2021, 22, 2941. [Google Scholar] [CrossRef]
- Ayadi, L.; Galvanin, A.; Pichot, F.; Marchand, V.; Motorin, Y. RNA Ribose Methylation (2′-O-Methylation): Occurrence, Biosynthesis and Biological Functions. Biochim. Biophys. Acta 2019, 1862, 253–269. [Google Scholar] [CrossRef]
- Wilkinson, M.L.; Crary, S.M.; Jackman, J.E.; Grayhack, E.J.; Phizicky, E.M. The 2′-O-Methyltransferase Responsible for Modification of Yeast TRNA at Position 4. RNA 2007, 13, 404–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprinzl, M.; Horn, C.; Brown, M.; Ioudovitch, A.; Steinberg, S. Compilation of TRNA Sequences and Sequences of TRNA Genes. Nucleic Acids Res. 1998, 26, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Marchand, V.; Motorin, Y.; Lafontaine, D.L.J. Identification of Sites of 2′-O-Methylation Vulnerability in Human Ribosomal RNAs by Systematic Mapping. Sci. Rep. 2017, 7, 11490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endres, L.; Rose, R.E.; Doyle, F.; Rahn, T.; Lee, B.; Seaman, J.; McIntyre, W.D.; Fabris, D. 2′-O-Ribose Methylation of Transfer RNA Promotes Recovery from Oxidative Stress in Saccharomyces Cerevisiae. PLoS ONE 2020, 15, e0229103. [Google Scholar] [CrossRef]
- Punekar, A.S.; Shepherd, T.R.; Liljeruhm, J.; Forster, A.C.; Selmer, M. Crystal Structure of RlmM, the 2′O-Ribose Methyltransferase for C2498 of Escherichia Coli 23S RRNA. Nucleic Acids Res. 2012, 40, 10507–10520. [Google Scholar] [CrossRef] [Green Version]
- Marcel, V.; Ghayad, S.E.; Belin, S.; Therizols, G.; Morel, A.-P.; Solano-Gonzàlez, E.; Vendrell, J.A.; Hacot, S.; Mertani, H.C.; Albaret, M.A.; et al. P53 Acts as a Safeguard of Translational Control by Regulating Fibrillarin and RRNA Methylation in Cancer. Cancer Cell 2013, 24, 318–330. [Google Scholar] [CrossRef] [Green Version]
- Shima, H.; Igarashi, K. N 1-Methyladenosine (M1A) RNA Modification: The Key to Ribosome Control. J. Biochem. 2020, 167, 535–539. [Google Scholar] [CrossRef]
- Jin, H.; Huo, C.; Zhou, T.; Xie, S. M1A RNA Modification in Gene Expression Regulation. Genes 2022, 13, 910. [Google Scholar] [CrossRef]
- Saikia, M.; Fu, Y.; Pavon-Eternod, M.; He, C.; Pan, T. Genome-Wide Analysis of N1-Methyl-Adenosine Modification in Human TRNAs. RNA 2010, 16, 1317–1327. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-Mediated TRNA Demethylation Regulates Translation. Cell 2016, 167, 816–828.e16. [Google Scholar] [CrossRef]
- Helm, M.; Giegé, R.; Florentz, C. A Watson-Crick Base-Pair-Disrupting Methyl Group (M1A9) Is Sufficient for Cloverleaf Folding of Human Mitochondrial TRNALys. Biochemistry 1999, 38, 13338–13346. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Hartmann, J.D.; Watzinger, P.; Klepper, A.; Peifer, C.; Kötter, P.; Lafontaine, D.L.J.; Entian, K.-D. A Single N1-Methyladenosine on the Large Ribosomal Subunit RRNA Impacts Locally Its Structure and the Translation of Key Metabolic Enzymes. Sci. Rep. 2018, 8, 11904. [Google Scholar] [CrossRef] [PubMed]
- Waku, T.; Nakajima, Y.; Yokoyama, W.; Nomura, N.; Kako, K.; Kobayashi, A.; Shimizu, T.; Fukamizu, A. NML-Mediated RRNA Base Methylation Links Ribosomal Subunit Formation to Cell Proliferation in a P53-Dependent Manner. J. Cell Sci. 2016, 129, 2382–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, S.; Suzuki, T.; Yokoyama, W.; Yagi, S.; Matsumura, K.; Nakajima, Y.; Harigae, H.; Fukamizu, A.; Motohashi, H. Nucleomethylin Deficiency Impairs Embryonic Erythropoiesis. J. Biochem. 2018, 163, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, X.; Wang, K.; Wang, L.; Shu, X.; Ma, S.; Yi, C. Transcriptome-Wide Mapping Reveals Reversible and Dynamic N(1)-Methyladenosine Methylome. Nat. Chem. Biol. 2016, 12, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, X.; Zhang, M.; Wang, K.; Chen, Y.; Zhou, J.; Mao, Y.; Lv, J.; Yi, D.; Chen, X.-W.; et al. Base-Resolution Mapping Reveals Distinct M1A Methylome in Nuclear- and Mitochondrial-Encoded Transcripts. Mol. Cell 2017, 68, 993–1005.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grozhik, A.V.; Olarerin-George, A.O.; Sindelar, M.; Li, X.; Gross, S.S.; Jaffrey, S.R. Antibody Cross-Reactivity Accounts for Widespread Appearance of M1A in 5′UTRs. Nat. Commun. 2019, 10, 5126. [Google Scholar] [CrossRef] [Green Version]
- Machnicka, M.A.; Olchowik, A.; Grosjean, H.; Bujnicki, J.M. Distribution and Frequencies of Post-Transcriptional Modifications in TRNAs. RNA Biol. 2014, 11, 1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Väre, V.Y.P.; Eruysal, E.R.; Narendran, A.; Sarachan, K.L.; Agris, P.F. Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules 2017, 7, 29. [Google Scholar] [CrossRef] [Green Version]
- Kato, T.; Daigo, Y.; Hayama, S.; Ishikawa, N.; Yamabuki, T.; Ito, T.; Miyamoto, M.; Kondo, S.; Nakamura, Y. A Novel Human TRNA-Dihydrouridine Synthase Involved in Pulmonary Carcinogenesis. Cancer Res. 2005, 65, 5638–5646. [Google Scholar] [CrossRef]
- Kowalski, S.; Yamane, T.; Fresco, J.R. Nucleotide Sequence of the “Denaturable” Leucine Transfer RNA from Yeast. Science 1971, 172, 385–387. [Google Scholar] [CrossRef]
- Kruppa, J.; Zachau, H.G. Multiplicity of Serine-Specific Transfer RNAs of Brewer’s and Baker’s Yeast. Biochim. Biophys. Acta 1972, 277, 499–512. [Google Scholar] [CrossRef]
- Kumbhar, B.V.; Kamble, A.D.; Sonawane, K.D. Conformational Preferences of Modified Nucleoside N(4)-Acetylcytidine, Ac4C Occur at “Wobble” 34th Position in the Anticodon Loop of TRNA. Cell Biochem. Biophys. 2013, 66, 797–816. [Google Scholar] [CrossRef]
- Sharma, S.; Langhendries, J.-L.; Watzinger, P.; Kötter, P.; Entian, K.-D.; Lafontaine, D.L.J. Yeast Kre33 and Human NAT10 Are Conserved 18S RRNA Cytosine Acetyltransferases That Modify TRNAs Assisted by the Adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015, 43, 2242–2258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominissini, D.; Rechavi, G. N4-Acetylation of Cytidine in MRNA by NAT10 Regulates Stability and Translation. Cell 2018, 175, 1725–1727. [Google Scholar] [CrossRef] [Green Version]
- Silva, A.P.G.; Byrne, R.T.; Chechik, M.; Smits, C.; Waterman, D.G.; Antson, A.A. Expression, Purification, Crystallization and Preliminary X-Ray Studies of the TAN1 Orthologue from Methanothermobacter Thermautotrophicus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 1083–1086. [Google Scholar] [CrossRef]
- Johansson, M.J.O.; Byström, A.S. The Saccharomyces Cerevisiae TAN1 Gene Is Required for N4-Acetylcytidine Formation in TRNA. RNA 2004, 10, 712–719. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Qin, Q.; Shi, X.; He, J.; Xu, G. Modified Metabolites Mapping by Liquid Chromatography-High Resolution Mass Spectrometry Using Full Scan/All Ion Fragmentation/Neutral Loss Acquisition. J. Chromatogr. A 2019, 1583, 80–87. [Google Scholar] [CrossRef]
- Arango, D.; Sturgill, D.; Oberdoerffer, S. Immunoprecipitation and Sequencing of Acetylated RNA. Bio-Protocol 2019, 9, e3278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Expanding the Nucleotide Repertoire of the Ribosome with Post-Transcriptional Modifications—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/17894445/ (accessed on 21 October 2022).
- Jenner, L.; Melnikov, S.; Garreau de Loubresse, N.; Ben-Shem, A.; Iskakova, M.; Urzhumtsev, A.; Meskauskas, A.; Dinman, J.; Yusupova, G.; Yusupov, M. Crystal Structure of the 80S Yeast Ribosome. Curr. Opin. Struct. Biol. 2012, 22, 759–767. [Google Scholar] [CrossRef]
- Rubio, M.A.T.; Gaston, K.W.; McKenney, K.M.; Fleming, I.M.C.; Paris, Z.; Limbach, P.A.; Alfonzo, J.D. Editing and Methylation at a Single Site by Functionally Interdependent Activities. Nature 2017, 542, 494–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basturea, G.N.; Rudd, K.E.; Deutscher, M.P. Identification and Characterization of RsmE, the Founding Member of a New RNA Base Methyltransferase Family. RNA 2006, 12, 426–434. [Google Scholar] [CrossRef] [Green Version]
- Yusupov, M.M.; Yusupova, G.Z.; Baucom, A.; Lieberman, K.; Earnest, T.N.; Cate, J.H.; Noller, H.F. Crystal Structure of the Ribosome at 5.5 A Resolution. Science 2001, 292, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Yang, C.-G.; Yang, S.; Jian, X.; Yi, C.; Zhou, Z.; He, C. Oxidative Demethylation of 3-Methylthymine and 3-Methyluracil in Single-Stranded DNA and RNA by Mouse and Human FTO. FEBS Lett. 2008, 582, 3313–3319. [Google Scholar] [CrossRef] [Green Version]
- Trixl, L.; Lusser, A. The Dynamic RNA Modification 5-Methylcytosine and Its Emerging Role as an Epitranscriptomic Mark. Wiley Interdiscip. Rev. RNA 2019, 10, e1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motorin, Y.; Lyko, F.; Helm, M. 5-Methylcytosine in RNA: Detection, Enzymatic Formation and Biological Functions. Nucleic Acids Res. 2010, 38, 1415–1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohnsack, K.E.; Höbartner, C.; Bohnsack, M.T. Eukaryotic 5-Methylcytosine (M5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes 2019, 10, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schosserer, M.; Minois, N.; Angerer, T.B.; Amring, M.; Dellago, H.; Harreither, E.; Calle-Perez, A.; Pircher, A.; Gerstl, M.P.; Pfeifenberger, S.; et al. Methylation of Ribosomal RNA by NSUN5 Is a Conserved Mechanism Modulating Organismal Lifespan. Nat. Commun. 2015, 6, 6158. [Google Scholar] [CrossRef] [Green Version]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread Occurrence of 5-Methylcytosine in Human Coding and Non-Coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef]
- Tuorto, F.; Herbst, F.; Alerasool, N.; Bender, S.; Popp, O.; Federico, G.; Reitter, S.; Liebers, R.; Stoecklin, G.; Gröne, H.-J.; et al. The TRNA Methyltransferase Dnmt2 Is Required for Accurate Polypeptide Synthesis during Haematopoiesis. EMBO J. 2015, 34, 2350–2362. [Google Scholar] [CrossRef]
- Blaze, J.; Navickas, A.; Phillips, H.L.; Heissel, S.; Plaza-Jennings, A.; Miglani, S.; Asgharian, H.; Foo, M.; Katanski, C.D.; Watkins, C.P.; et al. Neuronal Nsun2 Deficiency Produces TRNA Epitranscriptomic Alterations and Proteomic Shifts Impacting Synaptic Signaling and Behavior. Nat. Commun. 2021, 12, 4913. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Pan, K.; Fang, S.; Ye, L.; Tong, X.; Wang, Z.; Xue, X.; Zhang, H. Advances in MRNA 5-Methylcytosine Modifications: Detection, Effectors, Biological Functions, and Clinical Relevance. Mol. Ther. Nucleic Acids 2021, 26, 575–593. [Google Scholar] [CrossRef] [PubMed]
- Edelheit, S.; Schwartz, S.; Mumbach, M.R.; Wurtzel, O.; Sorek, R. Transcriptome-Wide Mapping of 5-Methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals M5C within Archaeal MRNAs. PLoS Genet. 2013, 9, e1003602. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.; Chen, W.; Liu, J.; Gu, N.; Zhang, R. Genome-Wide Identification of MRNA 5-Methylcytosine in Mammals. Nat. Struct. Mol. Biol. 2019, 26, 380–388. [Google Scholar] [CrossRef]
- Li, Q.; Li, X.; Tang, H.; Jiang, B.; Dou, Y.; Gorospe, M.; Wang, W. NSUN2-Mediated M5C Methylation and METTL3/METTL14-Mediated M6A Methylation Cooperatively Enhance P21 Translation. J. Cell. Biochem. 2017, 118, 2587–2598. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhang, Q.; Shi, Y.; Shi, Q.; Jiang, Y.; Gu, Y.; Li, Z.; Li, X.; Zhao, K.; Wang, C.; et al. Tet2 Promotes Pathogen Infection-Induced Myelopoiesis through MRNA Oxidation. Nature 2018, 554, 123–127. [Google Scholar] [CrossRef]
- Sun, Z.; Xue, S.; Zhang, M.; Xu, H.; Hu, X.; Chen, S.; Liu, Y.; Guo, M.; Cui, H. Aberrant NSUN2-Mediated M5C Modification of H19 LncRNA Is Associated with Poor Differentiation of Hepatocellular Carcinoma. Oncogene 2020, 39, 6906–6919. [Google Scholar] [CrossRef]
- Sajini, A.A.; Choudhury, N.R.; Wagner, R.E.; Bornelöv, S.; Selmi, T.; Spanos, C.; Dietmann, S.; Rappsilber, J.; Michlewski, G.; Frye, M. Loss of 5-Methylcytosine Alters the Biogenesis of Vault-Derived Small RNAs to Coordinate Epidermal Differentiation. Nat. Commun. 2019, 10, 2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Song, B.; Wei, Z.; Huang, D.; Zhang, Y.; Su, J.; de Magalhães, J.P.; Rigden, D.J.; Meng, J.; Chen, K. M5C-Atlas: A Comprehensive Database for Decoding and Annotating the 5-Methylcytosine (M5C) Epitranscriptome. Nucleic Acids Res. 2022, 50, D196–D203. [Google Scholar] [CrossRef]
- Ramanathan, A.; Robb, G.B.; Chan, S.-H. MRNA Capping: Biological Functions and Applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef]
- Haag, S.; Kretschmer, J.; Bohnsack, M.T. WBSCR22/Merm1 Is Required for Late Nuclear Pre-Ribosomal RNA Processing and Mediates N7-Methylation of G1639 in Human 18S rRNA. RNA 2015, 21, 180–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinther, J. No Evidence for N7-Methylation of Guanosine (M7G) in Human Let-7e. Mol. Cell 2020, 79, 199–200. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T.; Pandolfini, L.; Barbieri, I.; Bannister, A.J.; Andrews, B. Further Evidence Supporting N7-Methylation of Guanosine (M7G) in Human MicroRNAs. Mol. Cell 2020, 79, 201–202. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T. The Expanding World of TRNA Modifications and Their Disease Relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F.; Vendeix, F.A.P.; Graham, W.D. TRNA’s Wobble Decoding of the Genome: 40 Years of Modification. J. Mol. Biol. 2007, 366, 1–13. [Google Scholar] [CrossRef]
- Richner, J.M.; Himansu, S.; Dowd, K.A.; Butler, S.L.; Salazar, V.; Fox, J.M.; Julander, J.G.; Tang, W.W.; Shresta, S.; Pierson, T.C.; et al. Modified MRNA Vaccines Protect against Zika Virus Infection. Cell 2017, 168, 1114–1125.e10. [Google Scholar] [CrossRef] [Green Version]
- Chahal, J.S.; Khan, O.F.; Cooper, C.L.; McPartlan, J.S.; Tsosie, J.K.; Tilley, L.D.; Sidik, S.M.; Lourido, S.; Langer, R.; Bavari, S.; et al. Dendrimer-RNA Nanoparticles Generate Protective Immunity against Lethal Ebola, H1N1 Influenza, and Toxoplasma Gondii Challenges with a Single Dose. Proc. Natl. Acad. Sci. USA 2016, 113, E4133–E4142. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.-S.; Webby, R.J. An MRNA Vaccine for Influenza. Nat. Biotechnol. 2012, 30, 1202–1204. [Google Scholar] [CrossRef]
- Endres, L.; Fasullo, M.; Rose, R. TRNA Modification and Cancer: Potential for Therapeutic Prevention and Intervention. Future Med. Chem. 2019, 11, 885–900. [Google Scholar] [CrossRef]
- Zhang, X.; Su, H.; Chen, H.; Li, Q.; Liu, X.; Zhang, L.; Wu, W.K.K.; Chan, M.T.V.; Chen, H. RNA Modifications in Gastrointestinal Cancer: Current Status and Future Perspectives. Biomedicines 2022, 10, 1918. [Google Scholar] [CrossRef]

| Modification | Code | Base | Domains | RNA Classes | Location and Frequency | Function | Methods | Diseases | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| N6-methyladenosine | m6A | A | All | rRNA, tRNA, mRNA, miRNA, lncRNA, circRNA, snRNA | mRNA: enriched in 5′-UTRs, around stop codons, and in 3′ UTRs | mRNA: regulates splicing events, induces mRNA instability, increases translation efficiency; lncRNA: plays a role in lncRNA-mediated transcriptional repression | DRS, HPLC MeRIP-seq, MiCLIP | Osteoporosis, HCC, obesity | [21,22,23,24,25,26,27] |
| Inosine | I | A | All | tRNA, mRNA, miRNA | tRNA: p. 34, 37, 57; mRNA: dsRNA regions | tRNA: required for wobble codon recognition; mRNA: affects stability and localization, can change protein sequence | LC-MS/MS, RT-PCR, DRS, de novo RNA-seq | HCC; gastric, colorectal, esophageal, glioblastoma, and lung cancers | [28,29,30,31,32,33] |
| Pseudouridine | Ψ or Y | U | All | rRNA, tRNA, mRNA, snRNA, Mt-tRNA, scaRNA, snoRNA, miRNA, lincRNA | tRNA: p. 55; mRNA: 5′UTR, 3′UTR, CDS | rRNA: plays a role in ribosome assembly and translational fidelity; tRNA: increases stability | Pseudo-Seq, DRS, PSI-seq, CeU-Seq, Ψ-Seq, RBS-seq | Prostate, breast, and lung cancers; HCC | [34,35,36,37,38,39,40,41] |
| 7-methylguanosine | m7G | G | All | miRNA, rRNA, mRNA, tRNA, | rRNA: SSU G1639 (in humans); tRNA: p. 46; mRNA: 4522 clusters within 2318 mRNAs in 293T cells | tRNA: increases stability; mRNA: regulates translation | m7G-MeRIP-seq, m7G-seq, DRS, m7G-miCLIP-seq, m7G-MaP-seq | HCC, PAD, lung cancer | [35,42,43,44,45,46,47,48] |
| Dihydrouridine | D | U | All | tRNA, mRNA, snoRNA | tRNA: p. 16, 17, 20, 20a, 20b, 47; mRNA: 130 sites in 112 transcripts in S. cerevisiae; snoRNA: 48 sites in 23 snoRNA in S. cerevisiae | tRNA: destabilizes the structure; mRNA: affects splicing and translation | D-seq, Rho-seq, LC-MS | Lung cancer | [49,50,51,52,53] |
| N1-methyladenosine | m1A | A | All | rRNA, tRNA, mRNA, lncRNA | rRNA: p.1322 in 28S; tRNA: p. 9, 14, 16, 22, 57, 58; mRNA: rare | rRNA: ensures proper folding; tRNA: ensures stability and proper folding | ARM-seq, m1A-seq, m1A-quant-seq, m1A-ID-seq, m1A-seq-SS, m1A-seq-TGIR, m1A-MAP, m1A-IP-seq | HCC; cervical, pancreatic, breast, and ovarian cancers | [17,54,55,56,57,58,59] |
| Queuosine | Q | G | Bacteria,eukaryotes | tRNA | tRNATyr, tRNAHis, tRNAAsn, tRNAAsp: p. 34 (humans) | Impacts coding potential of tRNA, protects tRNA from ribonuclease cleavage | UHPLC-MS/MS, LC-MC | T-cell lymphoma, colon cancer | [60,61,62,63] |
| 5-methylcytidine | m5C | C | All | rRNA, tRNA, mRNA, eRNA, miRNA, lncRNA, viral RNA, vault RNA, snRNA, snoRNA | rRNA: m5C3761 and m5C4413 (well-conserved in eukaryotes); tRNA: variable loop, anticodon loop, and T stem; mRNA: enriched in 5′UTR | rRNA: ensures proper folding and translational fidelity; tRNA: ensures proper folding and stability, codon–anticodon interactions, and reading frame maintenance; mRNA: affects stability, translation rate; lncRNA: affects stability; vault RNA: required for biogenesis | m5C-RIP-seq, Aza-IP-seq, miCLIP-seq, TAWO-seq, RNA-BisSeq, RBS-seq, DRS, MeRIP-seq | ARID, DS, lactic acidosis, breast cancer, hypotonia /floppy baby syndrome, metabolism | [14,40,48,64,65,66,67] |
| 5-formylcytidine | f5C | C | All | tRNA | mt-tRNAMet: p.C34 (always) | Required for decoding of AUA codon in mitochondria | HPLC-MS | NA | [68,69] |
| 5-methyluridine | m5U | U | All | rRNA, tRNA | tRNA: p. 54 | Ensures fidelity of translation, folding, and stability of tRNA | FICC-Seq, iCLIP, miCLIP-seq | Breast cancer, systemic lupus erythematosus | [70,71,72,73] |
| 2′-O-methylation | Nm | All | All | rRNA, tRNA, mRNA, snRNA, snoRNA | mRNA: 5′UTR, 3′UTR, CDS; tRNA: p. 4 | Nm increases the thermodynamic stability of RNA:RNA base pairs and stabilizes A-form RNA duplexes | RiboMethSeq, 2D-TLC, Nm-seq, 2′OMe-seq, RTL-P, DRS, RibOxi-seq | Asthma, Alzheimer’s disease | [48,74,75,76,77,78,79] |
| 1-methylguanosine | m1G | G | All | tRNA, mRNA | tRNALeuCUN, tRNAProCCN, tRNAArgCGG: p. 37 (always); tRNA: p. 9 (always in S. cerevisiae); mRNA (0.00046% G in S. cerevisiae) | tRNA: prevents frameshifting; mRNA: reduces translation fidelity in a position- and codon-dependent manner | MR-FISH, LC-MS/MS, DRS | NA | [80,81,82,83,84,85,86] |
| 5-methylaminomethyl- 2-thiouridine | mnm5s2U | U | All | tRNA | tRNALys3UUU: p. 34 (always) | Required for decoding of the lysine codons AAA and AAG by tRNALys3UUU | LC-MS/MS | NA | [87,88,89] |
| Wybutosine | yW | G | Archaea, eukaryotes | tRNA | tRNAPhe: p. 37 (often) | Stabilizes codon–anticodon interactions | LC-MS/MS | NA | [90,91] |
| N4-acetylcytidine | ac4C | C | All | rRNA, tRNA, mRNA | tRNA: p. 12 (always); rRNA: in helix 34, 45; mRNA: enriched in 5′UTR and CDS (4250 sites in HeLa; not present in HEK293; 0.1% C in S. cerevisiae mRNA) | tRNA: increases translation efficiency and accuracy; mRNA: increases stability | acRIP-seq, ac4C-seq | NA | [92,93,94,95] |
| 3-methylcytidine | m3C | C | Eukaryotes | tRNA | tRNA: p. 32 (30–90%, depending on tRNA) | Unknown | AlkAniline-seq, HAC-seq, ARM-seq, DM-tRNA-seq, mim-tRNA-seq, hydro-tRNA-seq, LC-MS/MS, NAIL-MS | Several types of cancer | [57,96,97,98,99,100,101] |
| Archaeosine | G+ | G | Archaea | tRNA | tRNA: p. 15 (always), p.13 (in some species) | Increases thermal stability of tRNA | HPLC | NA | [102,103] |
| 5-methoxycarbonylmethyl- 2-thiouridine | mcm5s2U | U | Eukaryotes | tRNA | tRNALysUUU, tRNAGluUUC, tRNAGlnUUG: p. 34 (always) | Improves decoding efficiency of tRNA | HPLC-MS | NA | [88,104] |
| 2-thiocytidine | s2C | C | Bacteria, eukaryotes | tRNA | tRNAArg, tRNASer2GCU: p. 32 (always) | Restricts recognition of certain wobble codons | HPLC-MS | NA | [105,106] |
| N2,N2-dimethylguanosine | m2,2G | G | Archaea, eukaryotes | tRNA, mRNA | tRNA: p. 10, p. 26 (always); mRNA (0.00051% G in S. cerevisiae) | tRNA: plays a role in tRNA folding and prevents tRNA from adopting wrong conformation; mRNA: reduces translation fidelity in a position- and codon-dependent manner | PhOxi-seq, LC-MS/MS | NA | [82,107,108] |
| N6-isopentenyladenosine | i6A | A | Bacteria, eukaryotes | tRNA | tRNAUNN: p. A37 (always) | Increases translation fidelity and efficiency of cognate codons | PHA6 assay, HPLC, Sanger | NA | [109,110,111] |
| N6-threonylcarbamoyladenosine | t6A | A | All | tRNA | tRNAANN: p. A37 (always) | Facilitates codon–anticodon pairing and prevents frameshift during protein synthesis | LC-MS/MS | MERFF, neurodegeneration, diabetes | [110,111,112] |
| 3-methyluridine | m3U | U | All | rRNA, tRNA | rRNA: U1498 (always in E. coli 16S), U2634 U2843 (in S. cerevisiae 25S); tRNAThr: p.U32 (always in T. brucei) | Unknown | MR-FISH, RP-HPLC | NA | [85,113,114] |
| Wyosine | imG | G | Archaea | tRNA | tRNAPhe: p. 37 (often) | Stabilizes codon–anticodon interactions | HPLC-MS | NA | [115,116] |
| 1-methylinosine | m1I | I | Bacteria, archaea, eukaryotes | tRNA | tRNAAla: p. 37 (eukaryotes); tRNA: p. 57 (archaea) | Unknown | LC-MS/MS | NA | [83,117] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arzumanian, V.A.; Dolgalev, G.V.; Kurbatov, I.Y.; Kiseleva, O.I.; Poverennaya, E.V. Epitranscriptome: Review of Top 25 Most-Studied RNA Modifications. Int. J. Mol. Sci. 2022, 23, 13851. https://doi.org/10.3390/ijms232213851
Arzumanian VA, Dolgalev GV, Kurbatov IY, Kiseleva OI, Poverennaya EV. Epitranscriptome: Review of Top 25 Most-Studied RNA Modifications. International Journal of Molecular Sciences. 2022; 23(22):13851. https://doi.org/10.3390/ijms232213851
Chicago/Turabian StyleArzumanian, Viktoriia A., Georgii V. Dolgalev, Ilya Y. Kurbatov, Olga I. Kiseleva, and Ekaterina V. Poverennaya. 2022. "Epitranscriptome: Review of Top 25 Most-Studied RNA Modifications" International Journal of Molecular Sciences 23, no. 22: 13851. https://doi.org/10.3390/ijms232213851