Biomarkers of Periodontitis and Its Differential DNA Methylation and Gene Expression in Immune Cells: A Systematic Review
Abstract
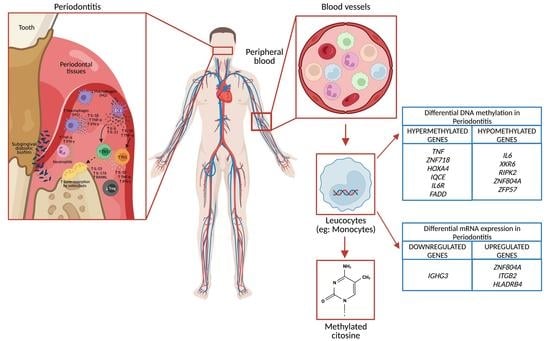
:1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
2.1.1. Inclusion Criteria
- Studies reporting potential systemic biomarkers of periodontitis based on differential DNA methylation and differential gene expression in peripheral immune cell types.
- Cross-sectional and case-control studies published since 2006 and up to July 2021 in the English language were considered.
2.1.2. Exclusion Criteria
2.2. Information Sources and Search Strategy
2.3. Data Selection and Extraction
2.4. Outcome Measures
2.5. Risk of Bias Assessment
2.6. Data Synthesis
3. Results
3.1. Data Selection
3.2. Description of the Studies
3.3. Risk of Bias Assessment
3.4. DNA Methylation in Peripheral Blood Leukocytes
3.5. Gene Expression in Polymorphonuclears Cells
3.6. Gene Expression in Peripheral Blood Mononuclear Cells (Lymphocytes and Monocytes)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Clin. Periodontol. 2018, 45, S162–S170. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.S.; Jepsen, S.; Jin, L.; Otomo-Corgel, J. Impact of the global burden of periodontal diseases on health, nutrition and wellbeing of mankind: A call for global action. J. Clin. Periodontol. 2017, 44, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Nazir, M.; Al-Ansari, A.; Al-Khalifa, K.; Alhareky, M.; Gaffar, B.; Almas, K. Global Prevalence of Periodontal Disease and Lack of Its Surveillance. Sci. World J. 2020, 2020, 2146160. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Lamont, R.J. Dancing with the Stars: How Choreographed Bacterial Interactions Dictate Nososymbiocity and Give Rise to Keystone Pathogens, Accessory Pathogens, and Pathobionts. Trends Microbiol. 2016, 24, 477–489. [Google Scholar] [CrossRef]
- Kinane, D.F.; Mark Bartold, P. Clinical relevance of the host responses of periodontitis. Periodontol. 2000 2007, 43, 278–293. [Google Scholar] [CrossRef]
- Van Dyke, T.E. Cellular and molecular susceptibility determinants for periodontitis. Periodontol. 2000 2007, 45, 10–13. [Google Scholar] [CrossRef]
- Lod, S.; Johansson, T.; Abrahamsson, K.; Larsson, L. The influence of epigenetics in relation to oral health. Int. J. Dent. Hyg. 2014, 12, 48–54. [Google Scholar] [CrossRef]
- Stylianou, E. Epigenetics of chronic inflammatory diseases. J. Inflamm. Res. 2018, 12, 1–14. [Google Scholar] [CrossRef]
- Patel, D.; Lee, Y.J.; Chauhan, B.; Sidhu, L.; Heck, D.; Duck, H. Epigenetics-Epidisease-Epidrug: A Key Context Folded inside of Periodontal Diseases. Enliven Dent. Periodont. 2019. [Google Scholar]
- Larsson, L. Current Concepts of Epigenetics and Its Role in Periodontitis. Curr. Oral Health Rep. 2017, 4, 286–293. [Google Scholar] [CrossRef]
- Almiñana-Pastor, P.J.; Boronat-Catalá, M.; Micó-Martinez, P.; Bellot-Arcís, C.; Lopez-Roldan, A.; Alpiste-Illueca, F.M. Epigenetics and periodontics: A systematic review. Med. Oral Patol. Oral Cir. Bucal 2019, 24, e659–e672. [Google Scholar] [CrossRef]
- Schulz, S.; Immel, U.D.; Just, L.; Schaller, H.G.; Gläser, C.; Reichert, S. Epigenetic characteristics in inflammatory candidate genes in aggressive periodontitis. Hum. Immunol. 2016, 77, 71–75. [Google Scholar] [CrossRef]
- Stefani, F.A.; Viana, M.B.; Dupim, A.C.; Brito, J.A.R.; Gomez, R.S.; da Costa, J.E.; Moreira, P.R. Expression, polymorphism and methylation pattern of interleukin-6 in periodontal tissues. Immunobiology 2013, 218, 1012–1017. [Google Scholar] [CrossRef]
- Yin, L.; Chung, W.O. Epigenetic regulation of human β-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunol. 2011, 4, 409–419. [Google Scholar] [CrossRef]
- de Faria Amormino, S.A.; Arão, T.C.; Saraiva, A.M.; Gomez, R.S.; Dutra, W.O.; da Costa, J.E.; de Fátima Correia Silva, J.; Moreira, P.R. Hypermethylation and low transcription of TLR2 gene in chronic periodontitis. Hum. Immunol. 2013, 74, 1231–1236. [Google Scholar] [CrossRef]
- Barros, S.P.; Hefni, E.; Nepomuceno, R.; Offenbacher, S.; North, K. Targeting epigenetic mechanisms in periodontal diseases. Periodontol. 2000 2018, 78, 174–184. [Google Scholar] [CrossRef]
- Hernández, H.G.; Hernández-Castañeda, A.A.; Pieschacón, M.P.; Arboleda, H. ZNF718, HOXA4, and ZFP57 are differentially methylated in periodontitis in comparison with periodontal health: Epigenome-wide DNA methylation pilot study. J. Periodontal Res. 2021, 56, 710–725. [Google Scholar] [CrossRef]
- Korte, D.L.; Kinney, J. Personalized medicine: An update of salivary biomarkers for periodontal diseases. Periodontol. 2000 2016, 70, 26–37. [Google Scholar] [CrossRef]
- Li, Q.S.; Vasanthakumar, A.; Davis, J.W.; Idler, K.B.; Nho, K.; Waring, J.F.; Saykin, A.J.; for the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Association of peripheral blood DNA methylation level with Alzheimer’s disease progression. Clin. Epigenetics 2021, 13, 191. [Google Scholar] [CrossRef]
- Hernández, H.G.; Sandoval-Hernández, A.G.; Garrido-Gil, P.; Labandeira-Garcia, J.L.; Zelaya, M.V.; Bayon, G.F.; Fernández, A.F.; Fraga, M.F.; Arboleda, G.; Arboleda, H. Alzheimer’s disease DNA methylome of pyramidal layers in frontal cortex: Laser-assisted microdissection study. Epigenomics 2018, 10, 1365–1382. [Google Scholar] [CrossRef]
- Liu, Y.; Aryee, M.J.; Padyukov, L.; Fallin, M.D.; Hesselberg, E.; Runarsson, A.; Reinius, L.; Acevedo, N.; Taub, M.; Ronninger, M.; et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 2013, 31, 142–147. [Google Scholar] [CrossRef]
- Bell, C.G.; Teschendorff, A.E.; Rakyan, V.K.; Maxwell, A.P.; Beck, S.; Savage, D.A. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med. Genom. 2010, 3, 33. [Google Scholar] [CrossRef]
- Tost, J. DNA Methylation: An Introduction to the Biology and the Disease-Associated Changes of a Promising Biomarker. Mol. Biotechnol. 2010, 44, 71–81. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, 1–9. [Google Scholar] [CrossRef]
- Kurushima, Y.; Tsai, P.C.; Castillo-Fernandez, J.; Couto Alves, A.; El-Sayed Moustafa, J.S.; Le Roy, C.; Spector, T.D.; Ide, M.; Hughes, F.J.; Small, K.S.; et al. Epigenetic findings in periodontitis in UK twins: A cross-sectional study. Clin. Epigenetics. 2019, 11, 27. [Google Scholar] [CrossRef]
- Shaddox, L.M.; Mullersman, A.F.; Huang, H.; Wallet, S.M.; Langaee, T.; Aukhil, I. Epigenetic regulation of inflammation in localized aggressive periodontitis. Clin. Epigenetics 2017, 9, 94. [Google Scholar] [CrossRef]
- Oliveira, N.F.P.; Damm, G.R.; Andia, D.C.; Salmon, C.; Nociti, F.H., Jr.; Line, S.R.P.; De Souza, A.P. DNA methylation status of the IL8 gene promoter in oral cells of smokers and non-smokers with chronic periodontitis. J. Clin. Periodontol. 2009, 36, 719–725. [Google Scholar] [CrossRef]
- Kojima, A.; Kobayashi, T.; Ito, S.; Murasawa, A.; Nakazono, K.; Yoshie, H. Tumor necrosis factor-alpha gene promoter methylation in Japanese adults with chronic periodontitis and rheumatoid arthritis. J. Periodontal Res. 2016, 51, 350–358. [Google Scholar] [CrossRef]
- Ishida, K.; Kobayashi, T.; Ito, S.; Komatsu, Y.; Yokoyama, T.; Okada, M.; Abe, A.; Murasawa, A.; Yoshie, H. Interleukin-6 Gene Promoter Methylation in Rheumatoid Arthritis and Chronic Periodontitis. J. Periodontol. 2012, 83, 917–925. [Google Scholar] [CrossRef]
- Wright, H.J.; Matthews, J.B.; Chapple, I.L.C.; Ling-Mountford, N.; Cooper, P.R. Periodontitis Associates with a Type 1 IFN Signature in Peripheral Blood Neutrophils. J. Immunol. 2008, 181, 5775–5784. [Google Scholar] [CrossRef]
- Iwata, T.; Kantarci, A.; Yagi, M.; Jackson, T.; Hasturk, H.; Kurihara, H.; Van Dyke, T.E. Ceruloplasmin Induces Polymorphonuclear Leukocyte Priming in Localized Aggressive Periodontitis. J. Periodontol. 2009, 80, 1300–1306. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, L.; Havemose-Poulsen, A.; Sønder, S.U.; Bendtzen, K.; Holmstrup, P. Blood cell gene expression profiling in subjects with aggressive periodontitis and chronic arthritis. J. Periodontol. 2008, 79, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, J.R.; Gröger, S.; Boedeker, R.-H.; Meyle, J. Expression and secretion levels of Th1 and Th2 cytokines in patients with aggressive periodontitis. Clin. Oral Investig. 2012, 16, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Z.; Maney, P.; Puri, J.; Zhou, Y.; Baddoo, M.; Strong, M.; Wang, Y.-P.; Flemington, E.; Deng, H.-W. RNA-sequencing study of peripheral blood monocytes in chronic periodontitis. Gene 2016, 581, 152–160. [Google Scholar] [CrossRef]
- Corbi, S.C.T.; de Vasconcellos, J.F.; Bastos, A.S.; Bussaneli, D.G.; da Silva, B.R.; Santos, R.A.; Takahashi, C.S.; de S. Rocha, C.; Carvalho, B.d.S.; Maurer-Morelli, C.V.; et al. Circulating lymphocytes and monocytes transcriptomic analysis of patients with type 2 diabetes mellitus, dyslipidemia and periodontitis. Sci. Rep. 2020, 10, 8145. [Google Scholar] [CrossRef]
- Gonçalves Fernandes, J.; Morford, L.A.; Harrison, P.L.; Kompotiati, T.; Huang, H.; Aukhil, I.; Wallet, S.M.; Macchion Shaddox, L. Dysregulation of genes and microRNAs in localized aggressive periodontitis. J. Clin. Periodontol. 2020, 47, 1317–1325. [Google Scholar] [CrossRef]
- Oseni, S.O.; Adebayo, O.; Adebayo, A.; Kwakye, A.; Pavlovic, M.; Asghar, W.; Hartmann, J.; Fields, G.B.; Kumi-Diaka, J. Integrative genomic and epigenomic analyses identified IRAK1 as a novel target for chronic inflammation-driven prostate tumorigenesis. bioRxiv 2021, 2021, 1–74. [Google Scholar] [CrossRef]
- Oseni, S.O. Role of Interleukin-1 Receptor-Associated Kinases in Chronic Inflammation and Prostate Tumorigenesis. Florida Atlantic University, Boca Raton, FL, USA, 2021. [Google Scholar]
- Wysocki, K.; Conley, Y.; Wenzel, S. Epigenome variation in severe asthma. Biol. Res. Nurs. 2015, 17, 263–269. [Google Scholar] [CrossRef]
- Laaksonen, D.E.; Niskanen, L.; Punnonen, K.; Nyyssönen, K.; Tuomainen, T.P.; Salonen, R.; Rauramaa, R.; Salonen, J.T. Sex hormones, inflammation and the metabolic syndrome: A population-based study. Eur. J. Endocrinol. 2003, 149, 601–608. [Google Scholar] [CrossRef]
- Arathimos, R.; Sharp, G.C.; Granell, R.; Tilling, K.; Relton, C.L. Associations of sex hormone-binding globulin and testosterone with genome-wide DNA methylation. BMC Genet. 2018, 19, 113. [Google Scholar] [CrossRef]
- Zhang, S.; Barros, S.P.; Moretti, A.J.; Yu, N.; Zhou, J.; Preisser, J.S.; Niculescu, M.D.; Offenbacher, S. Epigenetic regulation of TNFA expression in periodontal disease. J. Periodontol. 2013, 84, 1606–1616. [Google Scholar] [CrossRef]
- Shanmugam, M.K.; Sethi, G. Role of epigenetics in inflammation-associated diseases. Subcell. Biochem. 2013, 61, 627–657. [Google Scholar] [CrossRef]
- Hirst, M.; Marra, M.A. Epigenetics and human disease. Int. J. Biochem. Cell Biol. 2009, 41, 136–146. [Google Scholar] [CrossRef]

| Database | Keywords |
|---|---|
| Medline via PubMed | (“Periodontitis” [Mesh]) AND (“Humans” [Mesh]) AND (“Gene Expression” [Mesh] OR “Methylation” [Mesh] OR “DNA Methylation” [Mesh] OR “Transcriptome” [Mesh] OR “Oligonucleotide Array Sequence Analysis” [Mesh] OR “Sequence Analysis, DNA” [Mesh]) AND (“Neutrophils” [Mesh] OR “Leukocytes” [Mesh] OR “Blood Cells” [Mesh] OR “Leukocytes, Mononuclear” [Mesh] OR “Monocytes” [Mesh] OR “Granulocytes” [Mesh] OR “Eosinophils” [Mesh] OR “Lymphocytes” [Mesh] OR “B-Lymphocytes” [Mesh] OR “T-Lymphocytes” [Mesh]) |
| Web of Science | TS = (periodontitis AND “Gene Expression” OR “Methylation” OR “DNA Methylation” OR “Transcriptome” OR “Oligonucleotide Array Sequence Analysis” OR “Sequence Analysis, DNA” AND “Neutrophils” OR “Leukocytes” OR “Blood Cells”) TS = humans TI = periodontitis |
| Scopus | TITLE (periodontitis) AND humans ANDTITLE-ABSKEY (“Gene Expression” OR “Methylation” OR “DNA Methylation” OR “Transcriptome” OR “Oligonucleotide Array Sequence Analysis” OR “Sequence Analysis, DNA”) AND TITLE-ABS-KEY (“Neutrophils” OR “Leukocytes” OR “Blood Cells” OR “mononuclear Cells” OR monocytes OR granulocytes OR eosinophils OR lymphocytes OR “B cells” OR “T cells”) |
| Google Scholar | (intitle:”periodontal” OR intitle:”periodontitis”) AND (“gene expression” OR “DNA methylation” OR “transcriptome”) AND (“polymorphonuclear” OR “blood” OR “peripheral blood” OR “leucocytes” OR monocytes) AND (microarrays OR “microchip”) |
| Gene Expression Omnibus | periodontitis AND (“Gene Expression” OR “Methylation”) AND(“Neutrophils” OR “Leukocytes” OR “Blood Cells”) |
| Study/Year | Focus | Type of Study | Evaluated Genes | Nominal Condition(s) of Interest/Periodontitis Definition Criteria (Clinical Parameters) |
|---|---|---|---|---|
| Oliveira N.F.P. et al., 2009 [27] | DNA methylation status in the gene promoter of IL8 in blood leucocytes | Cross-sectional | IL8 | Periodontitis/At least three teeth exhibiting sites ≥ 5 mm CAL, in at least two different quadrants |
| Ishida K. et al., 2012 [29] | DNA methylation IL6 promoter in mononuclear cells | Cross-sectional | IL6 | Periodontitis or Rheumatoid Arthritis/Sites with probing depth (PD) ≥ 4 mm |
| Kojima A. et al., 2016 [28] | DNA methylation pattern of the TNF promoter in blood cells | Cross-sectional | TNF | Periodontitis or Rheumatoid Arthritis/Sites with probing depth (PD) ≥ 4 mm |
| Shaddox L.M. et al., 2017 [26] | DNA methylation promoter regions of genes involved in TLR in peripheral blood cells | Cross-sectional | CD14, FADD, HRAS, HSPA1A, HSPD1, IL6R, IRAK1, IRAK2, IRF1, IRF3, IRF8, MAP3K7, MYD88, PPARA, RIPK2, TBK1, TLR2, TLR5, TOLLIP, TRAF6, UBE2N, UBE2V1, EP_SEC, EP_DEC Selected after the screening: FADD, MAP3K7, MYD88, PPARA, IRAK1, RIPK2, and IL6R | Periodontitis/CAL ≥ 4 mm localized in at least two teeth (first molar) |
| Kurushima Y. et al., 2019 [25] | Epigenomic variation in peripheral whole blood using a twofold approach | Cross-sectional | Genome-wide analysis Loci-focused analysis: NIN, ABHD12B, WHAMM, KCNK1, DAB2IP, CLEC19A, TRA, TM9SF2P, GGTA2P, IFI16, RBMS3, C1QTNF7, TSNARE, HPVC1, SLC15A4, PKP2, SNRPN, IL8, CD44, CXCL1, IL6ST, CCR1, MMP7, MMP13, MMP3, TLR9, IL18, IFNB1, GLT6D1, IL1B, IL1RN, IL6, IL10, VDR, CD14, TLR4, MMP1 RNA-sequencing ZNF804A, VDR, IL6ST, TMCO6, IL1RN, CD44, IL1B, WHAMM, and CXCL1 | Self-reported periodontitis traits/ • “Have you ever had the condition of gum bleeding” • “Have you ever had the condition of gum decay or loose teeth” |
| Hernández H.G. et al., 2021 [17] | DNA methylation in peripheral leukocytes | Cross-sectional | Genome-wide analysis | Periodontitis/NR |
| Author/Year | Focus | Type of Study | Evaluated Genes | Nominal Condition(s) of Interest/Periodontitis Definition Criteria |
|---|---|---|---|---|
| Wright H.J. et al., 2008 [30] | To analyze the gene expression signature of hyperresponsive peripheral blood neutrophils from periodontitis patients | Cross-sectional | Genome-wide analysis | Periodontitis/At least two non-adjacent sites per quadrant exhibiting PPD ≥ 5 mm, with bleeding on probing, radiographic bone loss ≥ 30% and were not first molar or incisor sites. |
| Iwata T. et al., 2009 [31] | To evaluate ceruloplasmin expression and regulation in human PMNs from healthy donors and patients diagnosed with P. | Cross-sectional | CP | Periodontitis/CAL ≥ 4 mm localized in at least two teeth (first molar) |
| Author/Year | Focus | Type of Study | Evaluated Genes | Nominal Condition(s) of Interest/Periodontitis Definition Criteria |
|---|---|---|---|---|
| Sørensen L.K. et al., 2008 [32] | Differentially expressed candidate genes in PBMCs | Cross-sectional | Genome-wide analysis | Periodontitis/CAL ≥ 4 mm localized at least two teeth or CAL ≥ 4 mm at least three teeth. |
| Gonzales J.R. et al., 2012 [33] | Expression and production of IL-2, IFNG, IL-4 and IL-13 in CD4+ cells from peripheral blood | Cross-sectional |
Th1 and Th2 cytokines (IL2, IFNG, IL4 and IL13) | Periodontitis/PPD and CAL ≥ 5 mm on at least one interproximal site affecting at least three teeth other than the first molars and incisors. |
| Liu Y.-Z. et al., 2016 [34] | Functional genes and pathways at monocyte transcriptomic level. | Cross-sectional | Genome-wide analysis | Periodontitis/CAL ≥ 5 mm |
| Corbi S.C.T. et al., 2020 [35] | Gene expression signatures from circulating lymphocytes | Cross-sectional | Genome-wide analysis | Periodontitis alone or associated with dyslipidemia and diabetes mellitus type 2/PD ≥ 6 mm and CAL ≥ 4 mm in at least 4 non-adjacent teeth. |
| Gonçalves Fernandes J et al., 2020 [36] | Gene expression of key TLR pathway genes and miRNA regulators in unstimulated PBMCs | Cross-sectional | 84 genes from TLR pathway RT2 Profiler PCR Arrays 84 genes miRNA genes from miScript PCR Arrays Human Immunopathology | Periodontitis/At least 2 sites with CAL > 2 mm and radiographic bone loss on first molar or incisor. |
| Cross-Sectional Studies | Selection | Comparability | Outcomes | Total | Overall Quality Assessment | ||||
|---|---|---|---|---|---|---|---|---|---|
| Representativeness of the Sample | Sample Size | Non Respondents | Ascertainment of the Exposure | Assessment of Outcomes | Statistical Test | ||||
| Corbi S.C.T. et al., 2020 | ✯ | ✯ | - | ✯✯ | ✯✯ | ✯✯ | ✯ | 9✯ | Very Good |
| Kojima A. et al., 2016 | ✯ | ✯ | - | ✯✯ | ✯✯ | ✯✯ | ✯ | 9✯ | Very Good |
| Oliveira N.F.P. et al., 2009 | ✯ | ✯ | - | ✯✯ | ✯✯ | ✯✯ | ✯ | 9✯ | Very Good |
| Sørensen L.K. et al., 2008 | ✯ | ✯ | - | ✯✯ | ✯✯ | ✯✯ | ✯ | 9✯ | Very Good |
| Kurushima Y. et al., 2019 | ✯ | ✯ | - | ✯✯ | ✯✯ | ✯✯ | - | 8✯ | Good |
| Hernández H.G. et al., 2021 | ✯ | - | - | ✯✯ | ✯✯ | ✯✯ | ✯ | 8✯ | Good |
| Gonçalves Fernandes J et al., 2020 | ✯ | ✯ | - | ✯✯ | ✯ | ✯✯ | ✯ | 8✯ | Good |
| Ishida K et al., 2012 | ✯ | ✯ | - | ✯✯ | ✯ | ✯✯ | ✯ | 8✯ | Good |
| Wright HJ et al., 2008 | ✯ | ✯ | - | ✯✯ | ✯ | ✯✯ | ✯ | 8✯ | Good |
| Shaddox L.M. et al., 2017 | ✯ | ✯ | - | ✯ | ✯ | ✯✯ | ✯ | 7✯ | Good |
| Liu Y.-Z. et al., 2016 | ✯ | ✯ | - | ✯ | ✯ | ✯✯ | ✯ | 7✯ | Good |
| Gonzales J.R. et al., 2012 | ✯ | ✯ | - | ✯ | ✯ | ✯✯ | ✯ | 7✯ | Good |
| Iwata T et al., 2009 | ✯ | ✯ | - | - | ✯ | ✯✯ | ✯ | 6✯ | Satisfactory |
| Authors | Subject/ Population | Comparison | Expression Technique | Methylation Technique | Systemic Biomarkers Meth/mRNA |
|---|---|---|---|---|---|
| Oliveira N.F.P. et al., 2009 [27] | 13 smokers with periodontitis 13 non-smokers with periodontitis | 13 healthy (never smoked) control subjects (absence of CAL and no sites with probing depth > 3 mm) | NR | Methylation-specific PCR (MSP) | No DNA methylation/ Transcriptional expression biomarkers found for IL8 |
| Ishida K et al., 2012 [29] | 30 patients with RA and 30 patients with Periodontitis | 30 age-, sex-, and smoking status–balanced healthy controls | NR | Direct bisulfite sequencing | Hypometh IL6 |
| Kojima A. et al., 2016 [28] | 30 patients with periodontitis (only) 30 patients with RA/ Japanese adults | 30 race-matched healthy controls | NR | Direct bisulfite sequencing (Signal correction using ESME) | Hypermeth TNF |
| Shaddox L.M. et al., 2017 [26] | 20 periodontitis/ African American 5–25 years old (10 initial and 10 advanced stages of the disease) | 20 healthy unrelated controls | NR |
EpiTect Methyl II PCR Array Human Toll-Like Receptor Signaling Pathway Signature Panel (Pyrosequencing) | Early stages of the disease Hypermeth MAP3K7 Hypermeth MYD88 Hypermeth IL6R Hypermeth RIPK2 Hypermeth IRAK1BP1 Hypermeth PPARA Hypermeth FADD Advanced stages of the disease Hypometh RIPK2 Hypometh MAP3K7 (at positions 1 and 3) Hypometh MYD88 (at positions 1 and 5) Hypometh IRAK1BP1 (at positions 1 and 3) Hypometh PPARA (at position 2) |
| Kurushima Y, et al., 2019 [25] | Patients with self-reported periodontitis Positive gingival bleeding trait: 269 female subjects (from 528 female individuals) Positive for tooth mobility trait: 121 female subjects (from 492 female individuals) RNA-sequencing (384 subjects) - Positive gingival bleeding trait: 342 (from 384 female individuals) - Positive for tooth mobility trait: 335 (from 384 female individuals) | Subjects without self-reported periodontitis Positive gingival bleeding trait: 259 female subjects (from 528 female individuals) Positive for tooth mobility trait: 371 female subjects (from 492 female individuals) RNA-sequencing (384 subjects) Negative gingival bleeding trait: 42 (from 384 female individuals) Negative for tooth mobility trait: 49 (from 384 female individuals) | RNA-sequencing | The Infinium Human Methylation 450 BeadChip | DNA Methylation in blood Hypometh ZNF804A ‡ in gingival bleeding. Hypermeth IQCE in tooth mobility Hypometh XKR6 in tooth mobility VDR, IL6ST, TMCO6, IL1RN, CD44, IL1B, WHAMM, and CXCL1 † mRNA Expression in Blood ↑ mRNA ZNF804A‡ in gingival bleeding WHAMM, TMCO6 † |
| Hernández H.G. et al., 2021 [17] | 8 periodontitis patients | 8 periodontally healthy subjects | NR | Illumina MethylationEPIC BeadChip (IMEB) | Hypermeth ZNF718 Hypermeth HOXA4 Hypometh ZFP57 |
| Authors | Subject/ Population | Comparison | Cell Type(s)/ Source | Expression Analysis | Systemic Biomarkers mRNA |
|---|---|---|---|---|---|
| Wright HJ et al., 2008 [30] | 19 patients with periodontitis (19, 36–61 years) (baseline and 3 months after periodontal treatment) | 19 Age- and gender-matched periodontally healthy control subjects (37–62 years) | Neutrophils (discontinuous Percoll gradient isolation)/ peripheral blood |
HG_U133A microarrays (Affymetrix) semi-quantitative RT-PCR | ↑ mRNA MX1, IFIT4, G1P2, IFIT1, CIG5, and IFI44-like |
| Iwata T et al., 2009 [31] | 36 patients with periodontitis (age range: 16 to 41 years) | 36 systemically healthy control subjects (n = 36; age range: 21 to 39 years) | PMNs discontinuous gradient | RT-qPCR | ↑ mRNA CP |
| Authors | Subject/ Population | Comparison | Cell Type(s)/ Source | Expression Technique | Systemic Biomarkers mRNA |
|---|---|---|---|---|---|
| Sørensen L.K. et al., 2008 [32] | For microarray 5 Subjects with periodontally untreated: For RT-PCR 45 subjects with periodontitis | For microarray 2 controls with healthy periodontium (no interproximal attachment loss and no clinical signs of oral inflammatory conditions) For RT-PCR 25 healthy control subjects | Mononuclear cells (density centrifugation)/ peripheral blood | HG-U133A expression array RT-PCR, qPCR | Periodontitis ↑ mRNA MYOM2 ↑ mRNA TLR2 |
| Gonzales J.R. et al., 2012 [33] | 20 periodontitis | 20 non-periodontitis control subjects | CD4+ cells/peripheral blood | Real-time polymerase chain reaction (RT PCR) (TaqMan®) | In the inactivated CD4+ cells in periodontitis: ↓ mRNA IL4 |
| Liu Y.-Z. et al., 2016 [34] | 5 periodontitis subjects (non-smoking) | 5 periodontally healthy | Mononuclear cells/peripheral blood | RNA-seq (Illumina TruSeq) microarray dataset GSE6751 | Periodontitis pathogenesis ↑ mRNA FACR, CLCN5 CUX1, RNASE3, REL, VNN2, SGMS2, GGT1, HLA-DOA, ME1 ↓ mRNA PXN-AS1, URGCP, RPS20, FAM98A, XBP1, G3BP1, NFAT5 and ZNF207. Endocytosis ↑ mRNA DENND1A, RUFY1, CORO1C, ASGR2, APP, DAB2, PICALM, CD36, AP1S2, CLEC7A, THBS1, CLCN5 and RIN3 Cytokine production ↑ mRNA NLRC4, G6PD, MYD88, TLR4, NLRP3 and PTAFR Apoptosis ↑ mRNA ARHGEF2, SGK1, DNM1L, XIAP, UBE4B, CIDEB, STK17B, TRIO, NLRP3, BCL2L13, NCSTN, TNFRSF1A, PEA15, NLRC4, APP, GSN, HIPK3, BNIP3L, NLRP12, and THBS1 |
| Corbi S.C.T. et al., 2020 [35] | 24 periodontitis patients for Microarray expression (U133 Plus 2.0, Affimetrix): • 5 poorly controlled T2DM + dyslipidemia + periodontitis (T2DMpoorly-DL-P) • 7 well-controlled T2DM with dyslipidemia and periodontitis (T2DMwell-DL-P) • 6 well-controlled T2DM + dyslipidemia + periodontitis (DL-P) • 6 normoglycemic individuals + dyslipidemia + periodontitis (P) 120 periodontitis patients for RT-qPCR validation of selected DEGs: • 30 T2DMpoorly-DL-P • 30 T2DMwell-DL-P • 30 DL-P • 30 P | For Microarray U133 Plus 2.0: • 6 systemically healthy individuals without periodontitis (H) (homogeneity regarding biochemical, lipid and clinical periodontal parameters). For RT-qPCR validation: - 30 H | Mono-nuclear cells (Lymphocytes and monocytes)/peripheral blood | Expression Microarray U133 Plus 2.0 RT-qPCR (validation) | • P ↓ mRNA IGHG3 ↑ mRNA ITGB2 and HLADRB4. • T2DMpoorly + DL + P ↑ mRNA TGFB1I1, VNN1 ↓ mRNA HLADRB4 and CXCL8 • T2Dmwell + DL + P ↓ mRNA BPTF, PDE3B ↑ mRNA FN1 • DL + P ↑ mRNA DAB2 ↓ mRNACD47 and HLADRB4 |
| Gonçalves Fernandes J et al., 2020 [36] | For array screening. 10 subjects with periodontitis for mRNA screening RT2 Profiler PCR Arrays (TLR pathway) microRNA ARRAY • 11 subjects with periodontitis for microRNA screening by miScript Immunopathology PCR arrays (Qiagen) For gene expression validation: • 29 periodontally healthy subjects for mRNA(qPCR) • 31 periodontally healthy subjects for microRNAs (qPCR) African American subjects | For array screening. • 9 control subjects for mRNA screening RT2 Profiler PCR Arrays (TLR pathway) • 11 control subjects for microRNA screening by miScript Immunopathology PCR arrays (Qiagen) For gene expression validation: 29 periodontally healthy subjects for mRNA(qPCR) 31 periodontally healthy subjects for microRNAs (qPCR) | Mononuclear cells (SepMate™ Isolation method)/peripheral blood |
-RT2 Profiler PCR Arrays (TLR pathway) -miScript PCR Arrays Human Immunopathology -RT-qPCR (validation) | ↑ mRNA TLR2, TICAM-1 (TRIF), IRAK1, FOS, CCL2 ↑ mRNA miRNAs MIR9-1, MIR155, MIR203A, MIR147A, MIR182, MIR183 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cárdenas, A.M.; Ardila, L.J.; Vernal, R.; Melgar-Rodríguez, S.; Hernández, H.G. Biomarkers of Periodontitis and Its Differential DNA Methylation and Gene Expression in Immune Cells: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 12042. https://doi.org/10.3390/ijms231912042
Cárdenas AM, Ardila LJ, Vernal R, Melgar-Rodríguez S, Hernández HG. Biomarkers of Periodontitis and Its Differential DNA Methylation and Gene Expression in Immune Cells: A Systematic Review. International Journal of Molecular Sciences. 2022; 23(19):12042. https://doi.org/10.3390/ijms231912042
Chicago/Turabian StyleCárdenas, Angélica M., Laura J. Ardila, Rolando Vernal, Samanta Melgar-Rodríguez, and Hernán G. Hernández. 2022. "Biomarkers of Periodontitis and Its Differential DNA Methylation and Gene Expression in Immune Cells: A Systematic Review" International Journal of Molecular Sciences 23, no. 19: 12042. https://doi.org/10.3390/ijms231912042