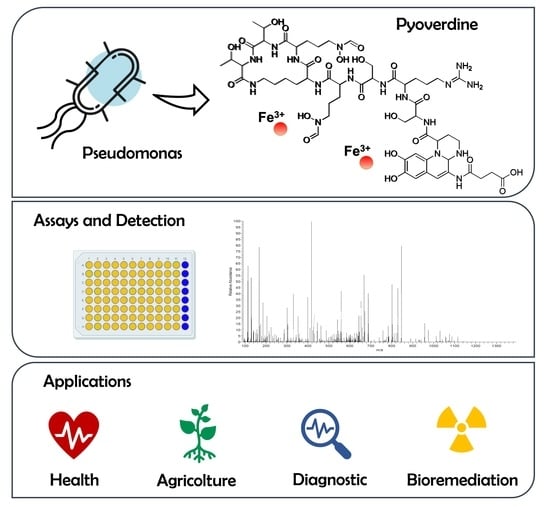
Novel Insights on Pyoverdine: From Biosynthesis to Biotechnological Application
Abstract
:1. Pyoverdines’ General Features
2. PVD Biosynthesis
3. Biological Functions of PVDs
3.1. Iron Uptake
3.2. PVDs’ Role in Pseudomonas Virulence
4. Inverting the Tide: PVDs as Target for Antimicrobial Therapy
5. Biotechnological Potential of PVDs
5.1. Applications
5.2. Cluster Engineering and Heterologous Expression of PVDs
6. Analytical Methods for the Identification of PVDs
6.1. Colorimetric and Electrochemical Detection
6.2. Mass Spectrometry and Structural Analysis
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gessard, M. Sur La Fonction Fluorescigène Des Microbes. Ann. Inst. Pasteur Paris 1892, 6, 801–823. [Google Scholar]
- Meyer, J.M.; Abdallah, M. The Fluorescentpigment OfPseudomonas Fluorescens: Biosynthesis, Puri-Fication and Physicochemical Properties. J. Gen. Microbiol. 1978, 107, 319–328. [Google Scholar] [CrossRef]
- Meyer, J.M.; Hornspreger, J. Role of Pyover-DinePfthe Iron Binding Fluorescent Pigment OfPseu-Domonas Fluorescensiron Transport. J. Gen. Microbiol. 1978, 107, 329–331. [Google Scholar] [CrossRef]
- Mular, A.; Shanzer, A.; Kozłowski, H.; Hubmann, I.; Misslinger, M.; Krzywik, J.; Decristoforo, C.; Gumienna-Kontecka, E. Cyclic Analogs of Desferrioxamine e Siderophore for 68Ga Nuclear Imaging: Coordination Chemistry and Biological Activity in Staphylococcus Aureus. Inorg. Chem. 2021, 60, 17846–17857. [Google Scholar] [CrossRef]
- Swayambhu, G.; Bruno, M.; Gulick, A.M.; Pfeifer, B.A. Siderophore Natural Products as Pharmaceutical Agents. Curr. Opin. Biotechnol. 2021, 69, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial Iron Homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Manck, L.E.; Park, J.; Tully, B.J.; Poire, A.M.; Bundy, R.M.; Dupont, C.L.; Barbeau, K.A. Petrobactin, a Siderophore Produced by Alteromonas, Mediates Community Iron Acquisition in the Global Ocean. ISME J. 2022, 16, 358–369. [Google Scholar] [CrossRef] [PubMed]
- GAUSE, G.F. Recent Studies on Albomycin, a New Antibiotic. Br. Med. J. 1955, 2, 1177–1179. [Google Scholar] [CrossRef] [PubMed]
- Brickman, T.J.; Hansel, J.-G.; Miller, M.J.; Armstrong, S.K. Purification, Spectroscopic Analysis and Biological Activity of the Macrocyclic Dihydroxamate Siderophore Alcaligin Produced by Bordetella Pertussis and Bordetella Bronchiseptica. Biometals 1996, 9, 191–203. [Google Scholar] [CrossRef]
- Takahashi, A.; Nakamura, H.; Kameyama, T.; Kurasawa, S.; Naganawa, H.; Okami, Y.; Takeuchi, T.; Umezawa, H.; Iitaka, Y. Bisucaberin, a New Siderophore, Sensitizing Tumor Cells to Macrophage-Mediated Cytolysis. II. Physico-Chemical Properties and Structure Determination. J. Antibiot. 1987, 40, 1671–1676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qi, L.; Chen, G.; Yin, W.-B. Discovery and Genetic Identification of Amphiphilic Coprogen Siderophores from Trichoderm Hypoxylon. Appl. Microbiol. Biotechnol. 2021, 105, 2831–2839. [Google Scholar] [CrossRef] [PubMed]
- Ijiri, M.; Fujiya, M.; Konishi, H.; Tanaka, H.; Ueno, N.; Kashima, S.; Moriichi, K.; Sasajima, J.; Ikuta, K.; Okumura, T. Ferrichrome Identified from Lactobacillus Casei ATCC334 Induces Apoptosis through Its Iron-Binding Site in Gastric Cancer Cells. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2017, 39, 1010428317711311. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Hurley, J.F.; Taylor, J.T.; Puckhaber, L.; Lehner, S.; Druzhinina, I.; Schumacher, R.; Kenerley, C.M. Ferricrocin, the Intracellular Siderophore of Trichoderma Virens, Is Involved in Growth, Conidiation, Gliotoxin Biosynthesis and Induction of Systemic Resistance in Maize. Biochem. Biophys. Res. Commun. 2018, 505, 606–611. [Google Scholar] [CrossRef]
- Roosenberg, J.M.; Miller, M.J. Total Synthesis of the Siderophore Danoxamine. J. Org. Chem. 2000, 65, 4833–4838. [Google Scholar] [CrossRef] [PubMed]
- Codd, R.; Richardson-Sanchez, T.; Telfer, T.J.; Gotsbacher, M.P. Advances in the Chemical Biology of Desferrioxamine B. ACS Chem. Biol. 2018, 13, 11–25. [Google Scholar] [CrossRef]
- Yamanaka, K.; Oikawa, H.; Ogawa, H.-O.; Hosono, K.; Shinmachi, F.; Takano, H.; Sakuda, S.; Beppu, T.; Ueda, K. Desferrioxamine E Produced by Streptomyces Griseus Stimulates Growth and Development of Streptomyces Tanashiensis. Microbiology 2005, 151, 2899–2905. [Google Scholar] [CrossRef]
- Zhai, C.; Summer, D.; Rangger, C.; Haas, H.; Haubner, R.; Decristoforo, C.; Fusarinine, C. A Novel Siderophore-Based Bifunctional Chelator for Radiolabeling with Gallium-68. J. Label. Compd. Radiopharm. 2015, 58, 209–214. [Google Scholar] [CrossRef]
- Sokol, P.A.; Darling, P.; Lewenza, S.; Corbett, C.R.; Kooi, C.D. Identification of a Siderophore Receptor Required for Ferric Ornibactin Uptake in Burkholderia Cepacia. Infect. Immun. 2000, 68, 6554–6560. [Google Scholar] [CrossRef]
- Müller, G.; Isowa, Y.; Raymond, K.N. Stereospecificity of Siderophore-Mediated Iron Uptake in Rhodotorula Pilimanae as Probed by Enantiorhodotorulic Acid and Isomers of Chromic Rhodotorulate. J. Biol. Chem. 1985, 260, 13921–13926. [Google Scholar] [CrossRef]
- Cornish, A.S.; Page, W.J. The Catecholate Siderophores of Azotobacter Vinelandii: Their Affinity for Iron and Role in Oxygen Stress Management. Microbiology 1998, 144 7, 1747–1754. [Google Scholar] [CrossRef]
- Dertz, E.A.; Stintzi, A.; Raymond, K.N. Siderophore-Mediated Iron Transport in Bacillus Subtilis and Corynebacterium Glutamicum. JBIC J. Biol. Inorg. Chem. 2006, 11, 1087–1097. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, I.G.; Cox, G.B.; Gibson, F. Biologically Active Compounds Containing 2,3-Duhydroxybenzoic Acid and Serine Formed by Escherichia Coli. Biochim. Biophys. Acta-Gen. Subj. 1970, 201, 453–460. [Google Scholar] [CrossRef]
- Wen, Y.; Wu, X.; Teng, Y.; Qian, C.; Zhan, Z.; Zhao, Y.; Li, O. Identification and Analysis of the Gene Cluster Involved in Biosynthesis of Paenibactin, a Catecholate Siderophore Produced by Paenibacillus Elgii B69. Environ. Microbiol. 2011, 13, 2726–2737. [Google Scholar] [CrossRef]
- Cornish, A.S.; Page, W.J. Production of the Triacetecholate Siderophore Protochelin by Azotobacter Vinelandii. Biometals 1995, 8, 332–338. [Google Scholar] [CrossRef]
- Bister, B.; Bischoff, D.; Nicholson, G.J.; Valdebenito, M.; Schneider, K.; Winkelmann, G.; Hantke, K.; Süssmuth, R.D. The Structure of Salmochelins: C-Glucosylated Enterobactins of Salmonella Enterica. BioMetals 2004, 17, 471–481. [Google Scholar] [CrossRef]
- Griffiths, G.L.; Sigel, S.P.; Payne, S.M.; Neilands, J.B. Vibriobactin, a Siderophore from Vibrio Cholerae. J. Biol. Chem. 1984, 259, 383–385. [Google Scholar] [CrossRef]
- Münzinger, M.; Budzikiewicz, H.; Expert, D.; Enard, C.; Meyer, J.M. Achromobactin, a New Citrate Siderophore of Erwinia Chrysanthemi. Z. Für Nat. C 2000, 55, 328–332. [Google Scholar] [CrossRef]
- Smith, M.J.; Neilands, J.B. Rhizobactin, a Siderophore from Rhizobium Meliloti. J. Plant Nutr. 1984, 7, 449–458. [Google Scholar] [CrossRef]
- Drechsel, H.; Tschierske, M.; Thieken, A.; Jung, G.; Zähner, H.; Winkelmann, G. The Carboxylate Type Siderophore Rhizoferrin and Its Analogs Produced by Directed Fermentation. J. Ind. Microbiol. 1995, 14, 105–112. [Google Scholar] [CrossRef]
- Konetschny-Rapp, S.; Jung, G.; Meiwes, J.; Zähner, H. Staphyloferrin A: A Structurally New Siderophore from Staphylococci. Eur. J. Biochem. 1990, 191, 65–74. [Google Scholar] [CrossRef]
- Montgomerie, J.Z.; Bindereif, A.; Neilands, J.B.; Kalmanson, G.M.; Guze, L.B. Association of Hydroxamate Siderophore (Aerobactin) with Escherichia Coli Isolated from Patients with Bacteremia. Infect. Immun. 1984, 46, 835–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seyedsayamdost, M.R.; Traxler, M.F.; Zheng, S.-L.; Kolter, R.; Clardy, J. Structure and Biosynthesis of Amychelin, an Unusual Mixed-Ligand Siderophore from Amycolatopsis Sp. AA4. J. Am. Chem. Soc. 2011, 133, 11434–11437. [Google Scholar] [CrossRef] [PubMed]
- Knosp, O.; Tigerstrom, M.; Page, W.J. Siderophore-Mediated Uptake of Iron in Azotobacter Vinelandii. J. Bacteriol. 1984, 159, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Unger, M.; Ntai, I.; McClure, R.A.; Albright, J.C.; Thomson, R.J.; Kelleher, N.L. Gobichelin A and B: Mixed-Ligand Siderophores Discovered Using Proteomics. Medchemcomm 2013, 4, 233–238. [Google Scholar] [CrossRef] [PubMed]
- De Voss, J.J.; Rutter, K.; Schroeder, B.G.; Su, H.; Zhu, Y.; Barry, C.E., 3rd. The Salicylate-Derived Mycobactin Siderophores of Mycobacterium Tuberculosis Are Essential for Growth in Macrophages. Proc. Natl. Acad. Sci. USA 2000, 97, 1252–1257. [Google Scholar] [CrossRef] [PubMed]
- Sonnenschein, E.C.; Stierhof, M.; Goralczyk, S.; Vabre, F.M.; Pellissier, L.; Hanssen, K.Ø.; de la Cruz, M.; Díaz, C.; de Witte, P.; Copmans, D.; et al. Pseudochelin A, a Siderophore of Pseudoalteromonas Piscicida S2040. Tetrahedron 2017, 73, 2633–2637. [Google Scholar] [CrossRef]
- Cox, C.D.; Adams, P. Siderophore Activity of Pyoverdin for Pseudomonas aeruginosa. Infect. Immun. 1985, 48, 130–138. [Google Scholar] [CrossRef]
- Dhungana, S.; Michalczyk, R.; Boukhalfa, H.; Lack, J.G.; Koppisch, A.T.; Fairlee, J.M.; Johnson, M.T.; Ruggiero, C.E.; John, S.G.; Cox, M.M.; et al. Purification and Characterization of Rhodobactin: A Mixed Ligand Siderophore from Rhodococcus Rhodochrous Strain OFS. BioMetals 2007, 20, 853–867. [Google Scholar] [CrossRef]
- Perry, R.D.; Balbo, P.B.; Jones, H.A.; Fetherston, J.D.; Demoll, E. Yersiniabactin from Yersinia Pestis: Biochemical Characterization of the Siderophore and Its Role in Iron Transport and Regulation. Microbiology 1999, 145, 1181–1190. [Google Scholar] [CrossRef]
- Sah, S.; Singh, R. Siderophore: Structural And Functional Characterisation—A Comprehensive Review. Agriculture 2015, 61, 97–114. [Google Scholar] [CrossRef]
- Gulick, A.M. Nonribosomal Peptide Synthetase Biosynthetic Clusters of ESKAPE Pathogens. Nat. Prod. Rep. 2017, 34, 981–1009. [Google Scholar] [CrossRef] [PubMed]
- Sayari, M.; van der Nest, M.A.; Steenkamp, E.T.; Soal, N.C.; Wilken, P.M.; Wingfield, B.D. Distribution and Evolution of Nonribosomal Peptide Synthetase Gene Clusters in the Ceratocystidaceae. Genes 2019, 10, 328. [Google Scholar] [CrossRef]
- Schalk, I.J.; Guillon, L. Pyoverdine Biosynthesis and Secretion in Pseudomonas aeruginosa: Implications for Metal Homeostasis. Environ. Microbiol. 2013, 15, 1661–1673. [Google Scholar] [CrossRef]
- D’Onofrio, A.; Crawford, J.M.; Stewart, E.J.; Witt, K.; Gavrish, E.; Epstein, S.; Clardy, J.; Lewis, K. Siderophores from Neighboring Organisms Promote the Growth of Uncultured Bacteria. Chem. Biol. 2010, 17, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Cordero, O.X.; Ventouras, L.-A.; DeLong, E.F.; Polz, M.F. Public Good Dynamics Drive Evolution of Iron Acquisition Strategies in Natural Bacterioplankton Populations. Proc. Natl. Acad. Sci. USA 2012, 109, 20059–20064. [Google Scholar] [CrossRef]
- Bonneau, A.; Roche, B.; Schalk, I.J. Iron Acquisition in Pseudomonas aeruginosa by the Siderophore Pyoverdine: An Intricate Interacting Network Including Periplasmic and Membrane Proteins. Sci. Rep. 2020, 10, 120. [Google Scholar] [CrossRef] [PubMed]
- Budzikiewicz, H.; Schäfer, M.; Fernández, D.U.; Matthijs, S.; Cornelis, P. Characterization of the Chromophores of Pyoverdins and Related Siderophores by Electrospray Tandem Mass Spectrometry. BioMetals 2007, 20, 135–144. [Google Scholar] [CrossRef]
- Ringel, M.T.; Brüser, T. The Biosynthesis of Pyoverdines. Microb. Cell 2018, 5, 424–437. [Google Scholar] [CrossRef]
- Albrecht-Gary, A.M.; Blanc, S.; Rochel, N.; Ocaktan, A.Z.; Abdallah, M.A. Bacterial Iron Transport: Coordination Properties of Pyoverdin PaA, a Peptidic Siderophore of Pseudomonas aeruginosa. Inorg. Chem. 1994, 33, 6391–6402. [Google Scholar] [CrossRef]
- Cézard, C.; Farvacques, N.; Sonnet, P. Chemistry and Biology of Pyoverdines, Pseudomonas Primary Siderophores. Curr. Med. Chem. 2015, 22, 165–186. [Google Scholar] [CrossRef]
- Meyer, J.M.; Neely, A.; Stintzi, A.; Georges, C.; Holder, I.A. Pyoverdin Is Essential for Virulence of Pseudomonas aeruginosa. Infect. Immun. 1996, 64, 518–523. [Google Scholar] [CrossRef] [Green Version]
- Visca, P.; Imperi, F.; Lamont, I.L. Pyoverdine Siderophores: From Biogenesis to Biosignificance. Trends Microbiol. 2007, 15, 22–30. [Google Scholar] [CrossRef]
- Gasser, V.; Guillon, L.; Cunrath, O.; Schalk, I.J. Cellular Organization of Siderophore Biosynthesis in Pseudomonas aeruginosa: Evidence for Siderosomes. J. Inorg. Biochem. 2015, 148, 27–34. [Google Scholar] [CrossRef]
- Ronnebaum, T.A.; Lamb, A.L. Nonribosomal Peptides for Iron Acquisition: Pyochelin Biosynthesis as a Case Study. Curr. Opin. Struct. Biol. 2018, 53, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Schalk, I.J.; Rigouin, C.; Godet, J. An Overview of Siderophore Biosynthesis among Fluorescent Pseudomonads and New Insights into Their Complex Cellular Organization. Environ. Microbiol. 2020, 22, 1447–1466. [Google Scholar] [CrossRef] [PubMed]
- Gross, H.; Loper, J.E. Genomics of Secondary Metabolite Production by Pseudomonas Spp. Nat. Prod. Rep. 2009, 26, 1408–1446. [Google Scholar] [CrossRef] [PubMed]
- Reimer, J.M.; Haque, A.S.; Tarry, M.J.; Schmeing, T.M. Piecing Together Nonribosomal Peptide Synthesis. Curr. Opin. Struct. Biol. 2018, 49, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Hannauer, M.; Schäfer, M.; Hoegy, F.; Gizzi, P.; Wehrung, P.; Mislin, G.L.A.; Budzikiewicz, H.; Schalk, I.J. Biosynthesis of the Pyoverdine Siderophore of Pseudomonas aeruginosa Involves Precursors with a Myristic or a Myristoleic Acid Chain. FEBS Lett. 2012, 586, 96–101. [Google Scholar] [CrossRef]
- Gasser, V.; Malrieu, M.; Forster, A.; Mély, Y.; Schalk, I.; Godet, J. Supra-Molecular Organization of the Pyoverdine Bio-Synthetic Pathway in Pseudomonas aeruginosa. bioRxiv 2018. [Google Scholar] [CrossRef]
- Durán, O.; Ramos, C.; Chen, O.; Castillo, J.; de Mayorga, B.; de Chial, M. Pyoverdine as an Important Virulence Factor in Pseudomonas aeruginosa Antibiotic Resistance. In The Global Antimicrobial Resistance Epidemic-Innovative Approaches and Cutting-Edge Solutions; Téllez, D.G., Ed.; IntechOpen: Rijeka, Croatia, 2022. [Google Scholar]
- Olucha, J.; Meneely, K.M.; Chilton, A.S.; Lamb, A.L. Two Structures of an N-Hydroxylating Flavoprotein Monooxygenase: Ornithine Hydroxylase from Pseudomonas aeruginosa. J. Biol. Chem. 2011, 286, 31789–31798. [Google Scholar] [CrossRef]
- Kenjić, N.; Hoag, M.R.; Moraski, G.C.; Caperelli, C.A.; Moran, G.R.; Lamb, A.L. PvdF of Pyoverdin Biosynthesis Is a Structurally Unique N(10)-Formyltetrahydrofolate-Dependent Formyltransferase. Arch. Biochem. Biophys. 2019, 664, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Gasser, V.; Malrieu, M.; Forster, A.; Mély, Y.; Schalk, I.J.; Godet, J. In Cellulo FRET-FLIM and Single Molecule Tracking Reveal the Supra-Molecular Organization of the Pyoverdine Bio-Synthetic Enzymes in Pseudomonas aeruginosa. Q. Rev. Biophys. 2020, 53, e1. [Google Scholar] [CrossRef] [PubMed]
- Philem, P.; Kleffmann, T.; Gai, S.; Hawkins, B.C.; Wilbanks, S.M.; Lamont, I.L. Identification of Active Site Residues of the Siderophore Synthesis Enzyme PvdF and Evidence for Interaction of PvdF with a Substrate-Providing Enzyme. Int. J. Mol. Sci. 2021, 22, 2211. [Google Scholar] [CrossRef]
- Winn, M.; Fyans, J.K.; Zhuo, Y.; Micklefield, J. Recent Advances in Engineering Nonribosomal Peptide Assembly Lines. Nat. Prod. Rep. 2016, 33, 317–347. [Google Scholar] [CrossRef]
- Baltz, R.H. Function of MbtH Homologs in Nonribosomal Peptide Biosynthesis and Applications in Secondary Metabolite Discovery. J. Ind. Microbiol. Biotechnol. 2011, 38, 1747–1760. [Google Scholar] [CrossRef]
- Lamont, I.L.; Martin, L.W.; Sims, T.; Scott, A.; Wallace, M. Characterization of a Gene Encoding an Acetylase Required for Pyoverdine Synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 3149–3152. [Google Scholar] [CrossRef]
- Greene, N.P.; Kaplan, E.; Crow, A.; Koronakis, V. Antibiotic Resistance Mediated by the MacB ABC Transporter Family: A Structural and Functional Perspective. Front. Microbiol. 2018, 9, 950. [Google Scholar] [CrossRef]
- Drake, E.J.; Gulick, A.M. Structural Characterization and High-Throughput Screening of Inhibitors of PvdQ, an NTN Hydrolase Involved in Pyoverdine Synthesis. ACS Chem. Biol. 2011, 6, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Nadal-Jimenez, P.; Koch, G.; Reis, C.R.; Muntendam, R.; Raj, H.; Jeronimus-Stratingh, C.M.; Cool, R.H.; Quax, W.J. PvdP Is a Tyrosinase That Drives Maturation of the Pyoverdine Chromophore in Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 2681–2690. [Google Scholar] [CrossRef]
- Ringel, M.T.; Dräger, G.; Brüser, T. PvdO Is Required for the Oxidation of Dihydropyoverdine as the Last Step of Fluorophore Formation in Pseudomonas Fluorescens. J. Biol. Chem. 2018, 293, 2330–2341. [Google Scholar] [CrossRef]
- Sugue, M.-F. Towards Understanding the Mechanisms of the Periplasmic Pyoverdine Maturation. Ph.D. Thesis, Institutionelles Repositorium der Leibniz Universität Hannover, Hannover, Germany, 2022. [Google Scholar]
- Ringel, M.T.; Dräger, G.; Brüser, T. PvdN Enzyme Catalyzes a Periplasmic Pyoverdine Modification. J. Biol. Chem. 2016, 291, 23929–23938. [Google Scholar] [CrossRef] [Green Version]
- Ringel, M.T.; Dräger, G.; Brüser, T. The Periplasmic Transaminase PtaA of Pseudomonas Fluorescens Converts the Glutamic Acid Residue at the Pyoverdine Fluorophore to α-Ketoglutaric Acid. J. Biol. Chem. 2017, 292, 18660–18671. [Google Scholar] [CrossRef] [PubMed]
- Henríquez, T.; Stein, N.V.; Jung, H. PvdRT-OpmQ and MdtABC-OpmB Efflux Systems Are Involved in Pyoverdine Secretion in Pseudomonas Putida KT2440. Environ. Microbiol. Rep. 2019, 11, 98–106. [Google Scholar] [CrossRef]
- Poole, K.; Neshat, S.; Krebes, K.; Heinrichs, D.E. Cloning and Nucleotide Sequence Analysis of the Ferripyoverdine Receptor Gene FpvA of Pseudomonas aeruginosa. J. Bacteriol. 1993, 175, 4597–4604. [Google Scholar] [CrossRef]
- Ghysels, B.; Dieu, B.T.M.; Beatson, S.A.; Pirnay, J.-P.; Ochsner, U.A.; Vasil, M.L.; Cornelis, P. FpvB, an Alternative Type I Ferripyoverdine Receptor of Pseudomonas aeruginosa. Microbiology 2004, 150, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Brillet, K.; Journet, L.; Célia, H.; Paulus, L.; Stahl, A.; Pattus, F.; Cobessi, D. A Beta Strand Lock Exchange for Signal Transduction in TonB-Dependent Transducers on the Basis of a Common Structural Motif. Structure 2007, 15, 1383–1391. [Google Scholar] [CrossRef]
- Brillet, K.; Ruffenach, F.; Adams, H.; Journet, L.; Gasser, V.; Hoegy, F.; Guillon, L.; Hannauer, M.; Page, A.; Schalk, I.J. An ABC Transporter with Two Periplasmic Binding Proteins Involved in Iron Acquisition in Pseudomonas aeruginosa. ACS Chem. Biol. 2012, 7, 2036–2045. [Google Scholar] [CrossRef]
- Schalk, I.J.; Abdallah, M.A.; Pattus, F. Recycling of Pyoverdin on the FpvA Receptor after Ferric Pyoverdin Uptake and Dissociation in Pseudomonas aeruginosa. Biochemistry 2002, 41, 1663–1671. [Google Scholar] [CrossRef]
- Greenwald, J.; Hoegy, F.; Nader, M.; Journet, L.; Mislin, G.L.A.; Graumann, P.L.; Schalk, I.J. Real Time Fluorescent Resonance Energy Transfer Visualization of Ferric Pyoverdine Uptake in Pseudomonas aeruginosa. A Role for Ferrous Iron. J. Biol. Chem. 2007, 282, 2987–2995. [Google Scholar] [CrossRef]
- Ganne, G.; Brillet, K.; Basta, B.; Roche, B.; Hoegy, F.; Gasser, V.; Schalk, I.J. Iron Release from the Siderophore Pyoverdine in Pseudomonas aeruginosa Involves Three New Actors: FpvC, FpvG, and FpvH. ACS Chem. Biol. 2017, 12, 1056–1065. [Google Scholar] [CrossRef]
- Vigouroux, A.; Aumont-Nicaise, M.; Boussac, A.; Marty, L.; Lo Bello, L.; Legrand, P.; Brillet, K.; Schalk, I.J.; Moréra, S. A Unique Ferrous Iron Binding Mode Is Associated with Large Conformational Changes for the Transport Protein FpvC of Pseudomonas aeruginosa. FEBS J. 2020, 287, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Imperi, F.; Tiburzi, F.; Visca, P. Molecular Basis of Pyoverdine Siderophore Recycling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2009, 106, 20440–20445. [Google Scholar] [CrossRef] [PubMed]
- Yeterian, E.; Martin, L.W.; Lamont, I.L.; Schalk, I.J. An Efflux Pump Is Required for Siderophore Recycling by Pseudomonas aeruginosa. Environ. Microbiol. Rep. 2010, 2 3, 412–418. [Google Scholar] [CrossRef]
- Jiricny, N.; Diggle, S.P.; West, S.A.; Evans, B.A.; Ballantyne, G.; Ross-Gillespie, A.; Griffin, A.S. Fitness Correlates with the Extent of Cheating in a Bacterium. J. Evol. Biol. 2010, 23, 738–747. [Google Scholar] [CrossRef]
- Butaitė, E.; Baumgartner, M.; Wyder, S.; Kümmerli, R. Siderophore Cheating and Cheating Resistance Shape Competition for Iron in Soil and Freshwater Pseudomonas Communities. Nat. Commun. 2017, 8, 414. [Google Scholar] [CrossRef] [PubMed]
- Butaitė, E.; Kramer, J.; Wyder, S.; Kümmerli, R. Environmental Determinants of Pyoverdine Production, Exploitation and Competition in Natural Pseudomonas Communities. Environ. Microbiol. 2018, 20, 3629–3642. [Google Scholar] [CrossRef]
- Tiburzi, F.; Imperi, F.; Visca, P. Intracellular Levels and Activity of PvdS, the Major Iron Starvation Sigma Factor of Pseudomonas aeruginosa. Mol. Microbiol. 2008, 67, 213–227. [Google Scholar] [CrossRef]
- Kümmerli, R.; Gardner, A.; West, S.A.; Griffin, A.S. Limited Dispersal, Budding Dispersal, and Cooperation: An Experimental Study. Evolution 2009, 63, 939–949. [Google Scholar] [CrossRef]
- Dumas, Z.; Ross-Gillespie, A.; Kümmerli, R. Switching between Apparently Redundant Iron-Uptake Mechanisms Benefits Bacteria in Changeable Environments. Proc. R. Soc. B Biol. Sci. 2013, 280, 20131055. [Google Scholar] [CrossRef]
- Loper, J.; Henkels, M. Availability of Iron to Pseudomonas Fluorescens in Rhizosphere and Bulk Soil Evaluated with an Ice Nucleation Reporter Gene. Appl. Environ. Microbiol. 1997, 63, 99–105. [Google Scholar] [CrossRef]
- Schalk, I.J. Metal Trafficking via Siderophores in Gram-Negative Bacteria: Specificities and Characteristics of the Pyoverdine Pathway. J. Inorg. Biochem. 2008, 102, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Kisaalita, W.S. Fluorescent Pseudomonad Pyoverdines Bind and Oxidize Ferrous Ion. Appl. Environ. Microbiol. 1998, 64, 1472–1476. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Turner, K.E.; Kirienko, N.V. PqsA Promotes Pyoverdine Production via Biofilm Formation. Pathogens 2017, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Kirienko, N. V High-Throughput Genetic Screen Reveals That Early Attachment and Biofilm Formation Are Necessary for Full Pyoverdine Production by Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 1707. [Google Scholar] [CrossRef]
- Palma, M.; DeLuca, D.; Worgall, S.; Quadri, L.E.N. Transcriptome Analysis of the Response of Pseudomonas aeruginosa to Hydrogen Peroxide. J. Bacteriol. 2004, 186, 248–252. [Google Scholar] [CrossRef]
- Dietrich, L.E.P.; Price-Whelan, A.; Petersen, A.; Whiteley, M.; Newman, D.K. The Phenazine Pyocyanin Is a Terminal Signalling Factor in the Quorum Sensing Network of Pseudomonas aeruginosa. Mol. Microbiol. 2006, 61, 1308–1321. [Google Scholar] [CrossRef]
- Sass, G.; Nazik, H.; Penner, J.; Shah, H.; Ansari, S.R.; Clemons, K.V.; Groleau, M.C.; Dietl, A.M.; Visca, P.; Haas, H.; et al. Aspergillus-Pseudomonas Interaction, Relevant to Competition in Airways. Med. Mycol. 2019, 57, S228–S232. [Google Scholar] [CrossRef]
- Sambrano, H.; Castillo, J.C.; Ramos, C.W.; de Mayorga, B.; Chen, O.; Durán, O.; Ciniglio, C.; Aguilar, C.; Cisterna, O.; de Chial, M. Prevalence of Antibiotic Resistance and Virulent Factors in Nosocomial Clinical Isolates of Pseudomonas aeruginosa from Panamá. Braz. J. Infect. Dis. 2020, 25, 101038. [Google Scholar] [CrossRef]
- Ullah, W.; Qasim, M.; Rahman, H.; Jie, Y.; Muhammad, N. Beta-lactamase-producing Pseudomonas aeruginosa: Phenotypic Characteristics and Molecular Identification of Virulence Genes. J. Chin. Med. Assoc. 2017, 80, 173–177. [Google Scholar] [CrossRef]
- Kang, D.; Kirienko, N.V. An in Vitro Cell Culture Model for Pyoverdine-Mediated Virulence. Pathogens 2021, 10, 9. [Google Scholar] [CrossRef]
- Bradshaw, J.L.; Caballero, A.R.; Bierdeman, M.A.; Adams, K.V.; Pipkins, H.R.; Tang, A.; O’Callaghan, R.J.; McDaniel, L.S. Pseudomonas aeruginosa Protease IV Exacerbates Pneumococcal Pneumonia and Systemic Disease. mSphere 2018, 3, e00212-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malloy, J.L.; Veldhuizen, R.A.W.; Thibodeaux, B.A.; O’Callaghan, R.J.; Wright, J.R. Pseudomonas aeruginosa Protease IV Degrades Surfactant Proteins and Inhibits Surfactant Host Defense and Biophysical Functions. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005, 288, L409–L418. [Google Scholar] [CrossRef] [PubMed]
- Guillon, A.; Brea, D.; Morello, E.; Tang, A.; Jouan, Y.; Ramphal, R.; Korkmaz, B.; Perez-Cruz, M.; Trottein, F.; O’Callaghan, R.J.; et al. Pseudomonas aeruginosa Proteolytically Alters the Interleukin 22-Dependent Lung Mucosal Defense. Virulence 2017, 8, 810–820. [Google Scholar] [CrossRef]
- Taguchi, F.; Suzuki, T.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. The Siderophore Pyoverdine of Pseudomonas Syringae Pv. Tabaci 6605 Is an Intrinsic Virulence Factor in Host Tobacco Infection. J. Bacteriol. 2010, 192, 117–126. [Google Scholar] [CrossRef]
- Kirienko, N.V.; Kirienko, D.R.; Larkins-Ford, J.; Wählby, C.; Ruvkun, G.; Ausubel, F.M. Pseudomonas aeruginosa Disrupts Caenorhabditis Elegans Iron Homeostasis, Causing a Hypoxic Response and Death. Cell Host Microbe 2013, 13, 406–416. [Google Scholar] [CrossRef]
- Kirienko, N.V.; Ausubel, F.M.; Ruvkun, G. Mitophagy Confers Resistance to Siderophore-Mediated Killing by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2015, 112, 1821–1826. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Kirienko, D.R.; Webster, P.; Fisher, A.L.; Kirienko, N.V. Pyoverdine, a Siderophore from Pseudomonas aeruginosa, Translocates into C. Elegans, Removes Iron, and Activates a Distinct Host Response. Virulence 2018, 9, 804–817. [Google Scholar] [CrossRef]
- Weigert, M.; Ross-Gillespie, A.; Leinweber, A.; Pessi, G.; Brown, S.P.; Kümmerli, R. Manipulating Virulence Factor Availability Can Have Complex Consequences for Infections. Evol. Appl. 2017, 10, 91–101. [Google Scholar] [CrossRef]
- Kirienko, D.R.; Kang, D.; Kirienko, N.V. Novel Pyoverdine Inhibitors Mitigate Pseudomonas aeruginosa Pathogenesis. Front. Microbiol. 2019, 10, 3317. [Google Scholar] [CrossRef]
- Rasko, D.A.; Sperandio, V. Anti-Virulence Strategies to Combat Bacteria-Mediated Disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef]
- Wang, X.; Kleerekoper, Q.; Revtovich, A.V.; Kang, D.; Kirienko, N.V. Identification and Validation of a Novel Anti-Virulent That Binds to Pyoverdine and Inhibits Its Function. Virulence 2020, 11, 1293–1309. [Google Scholar] [CrossRef] [PubMed]
- Rezzoagli, C.; Archetti, M.; Mignot, I.; Baumgartner, M.; Kümmerli, R. Combining Antibiotics with Antivirulence Compounds Can Have Synergistic Effects and Reverse Selection for Antibiotic Resistance in Pseudomonas aeruginosa. PLoS Biol. 2020, 18, e3000805. [Google Scholar] [CrossRef] [PubMed]
- Nicolafrancesco, C.; Porcaro, F.; Pis, I.; Nappini, S.; Simonelli, L.; Marini, C.; Frangipani, E.; Visaggio, D.; Visca, P.; Mobilio, S.; et al. Gallium- and Iron-Pyoverdine Coordination Compounds Investigated by X-ray Photoelectron Spectroscopy and X-ray Absorption Spectroscopy. Inorg. Chem. 2019, 58, 4935–4944. [Google Scholar] [CrossRef] [PubMed]
- Bonchi, C.; Frangipani, E.; Imperi, F.; Visca, P. Pyoverdine and Proteases Affect the Response of Pseudomonas aeruginosa to Gallium in Human Serum. Antimicrob. Agents Chemother. 2015, 59, 5641–5646. [Google Scholar] [CrossRef] [PubMed]
- Antonietti, V.; Boudesocque, S.; Dupont, L.; Farvacques, N.; Cézard, C.; Da Nascimento, S.; Raimbert, J.-F.; Socrier, L.; Robin, T.-J.; Morandat, S.; et al. Synthesis, Iron(III) Complexation Properties, Molecular Dynamics Simulations and P. Aeruginosa Siderophore-like Activity of Two Pyoverdine Analogs. Eur. J. Med. Chem. 2017, 137, 338–350. [Google Scholar] [CrossRef]
- Imperi, F.; Fiscarelli, E.V.; Visaggio, D.; Leoni, L.; Visca, P. Activity and Impact on Resistance Development of Two Antivirulence Fluoropyrimidine Drugs in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2019, 9, 49. [Google Scholar] [CrossRef]
- Rampioni, G.; Visca, P.; Leoni, L.; Imperi, F. Drug Repurposing for Antivirulence Therapy against Opportunistic Bacterial Pathogens. Emerg. Top. life Sci. 2017, 1, 13–22. [Google Scholar] [CrossRef]
- Kirienko, D.R.; Revtovich, A.V.; Kirienko, N.V. A High-Content, Phenotypic Screen Identifies Fluorouridine as an Inhibitor of Pyoverdine Biosynthesis and Pseudomonas aeruginosa Virulence. mSphere 2016, 1, e00217-16. [Google Scholar] [CrossRef] [PubMed]
- Rosy, J.C.; Babkiewicz, E.; Maszczyk, P.; Mrówka, P.; Kumar, B.K.; Murugesan, S.; Kunjiappan, S.; Sundar, K. L-Ornithine-N5-Monooxygenase (PvdA) Substrate Analogue Inhibitors for Pseudomonas aeruginosa Infections Treatment: Drug Repurposing Computational Studies. Biomolecules 2022, 12, 887. [Google Scholar] [CrossRef] [PubMed]
- Maenaka, R.; Tani, S.; Hikichi, Y.; Kai, K. Actinomycins Inhibit the Production of the Siderophore Pyoverdines in the Plant Pathogen Pseudomonas Cichorii SPC9018. Biosci. Biotechnol. Biochem. 2020, 84, 1975–1985. [Google Scholar] [CrossRef] [PubMed]
- Musthafa, K.S.; Voravuthikunchai, S.P. Anti-Virulence Potential of Eugenyl Acetate against Pathogenic Bacteria of Medical Importance. Antonie Van Leeuwenhoek 2015, 107, 703–710. [Google Scholar] [CrossRef]
- Bohac, T.J.; Fang, L.; Banas, V.S.; Giblin, D.E.; Wencewicz, T.A. Synthetic Mimics of Native Siderophores Disrupt Iron Trafficking in Acinetobacter Baumannii. ACS Infect. Dis. 2021, 7, 2138–2151. [Google Scholar] [CrossRef] [PubMed]
- Wibowo, J.P.; Batista, F.A.; van Oosterwijk, N.; Groves, M.R.; Dekker, F.J.; Quax, W.J. A Novel Mechanism of Inhibition by Phenylthiourea on PvdP, a Tyrosinase Synthesizing Pyoverdine of Pseudomonas aeruginosa. Int. J. Biol. Macromol. 2020, 146, 212–221. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, H.; Yu, H.; Dai, Q.; Xiong, J.; Sheng, H.; Qiu, J.; Jiang, L.; Peng, J.; He, X.; et al. Allicin Inhibits Pseudomonas aeruginosa Virulence by Suppressing the Rhl and Pqs Quorum-Sensing Systems. Can. J. Microbiol. 2019, 65, 563–574. [Google Scholar] [CrossRef]
- Neville, N.; Roberge, N.; Ji, X.; Stephen, P.; Lu, J.L.; Jia, Z. A Dual-Specificity Inhibitor Targets Polyphosphate Kinase 1 and 2 Enzymes To Attenuate Virulence of Pseudomonas aeruginosa. MBio 2021, 12, e0059221. [Google Scholar] [CrossRef]
- Petrik, M.; Umlaufova, E.; Raclavsky, V.; Palyzova, A.; Havlicek, V.; Haas, H.; Novy, Z.; Dolezal, D.; Hajduch, M.; Decristoforo, C. Imaging of Pseudomonas aeruginosa Infection with Ga-68 Labelled Pyoverdine for Positron Emission Tomography. Sci. Rep. 2018, 8, 15698. [Google Scholar] [CrossRef]
- Kinzel, O.; Tappe, R.; Gerus, I.; Budzikiewicz, H. The Synthesis and Antibacterial Activity of Two Pyoverdin-Ampicillin Conjugates, Entering Pseudomonas aeruginosa via the Pyoverdin-Mediated Iron Uptake Pathway. J. Antibiot. 1998, 51, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Kinzel, O.; Budzikiewicz, H. Synthesis and Biological Evaluation of a Pyoverdin-Beta-Lactam Conjugate: A New Type of Arginine-Specific Cross-Linking in Aqueous Solution. J. Pept. Res. 1999, 53, 618–625. [Google Scholar] [CrossRef]
- Hennard, C.; Truong, Q.C.; Desnottes, J.F.; Paris, J.M.; Moreau, N.J.; Abdallah, M.A. Synthesis and Activities of Pyoverdin-Quinolone Adducts: A Prospective Approach to a Specific Therapy against Pseudomonas aeruginosa. J. Med. Chem. 2001, 44, 2139–2151. [Google Scholar] [CrossRef]
- Mislin, G.L.A.; Schalk, I.J. Siderophore-Dependent Iron Uptake Systems as Gates for Antibiotic Trojan Horse Strategies against Pseudomonas aeruginosa. Metallomics 2014, 6, 408–420. [Google Scholar] [CrossRef] [PubMed]
- González, J.; Salvador, M.; Özkaya, Ö.; Spick, M.; Reid, K.; Costa, C.; Bailey, M.J.; Avignone Rossa, C.; Kümmerli, R.; Jiménez, J.I. Loss of a Pyoverdine Secondary Receptor in Pseudomonas aeruginosa Results in a Fitter Strain Suitable for Population Invasion. ISME J. 2021, 15, 1330–1343. [Google Scholar] [CrossRef] [PubMed]
- Ciui, B.; Tertiş, M.; Cernat, A.; Săndulescu, R.; Wang, J.; Cristea, C. Finger-Based Printed Sensors Integrated on a Glove for On-Site Screening Of Pseudomonas aeruginosa Virulence Factors. Anal. Chem. 2018, 90, 7761–7768. [Google Scholar] [CrossRef] [PubMed]
- Ferret, C.; Cornu, J.Y.; Elhabiri, M.; Sterckeman, T.; Braud, A.; Jezequel, K.; Lollier, M.; Lebeau, T.; Schalk, I.J.; Geoffroy, V.A. Effect of Pyoverdine Supply on Cadmium and Nickel Complexation and Phytoavailability in Hydroponics. Environ. Sci. Pollut. Res. Int. 2015, 22, 2106–2116. [Google Scholar] [CrossRef]
- Lee, E.T.; Lim, S.K.; Nam, D.H.; Khang, Y.H.; Kim, S.D. Pyoverdin2112 of Pseudomonas Fluorescens 2112 Inhibits Phytophthora Capsici, a Red-Pepper Blight-Causing Fungus. J. Microbiol. Biotechnol. 2003, 13, 415–421. [Google Scholar]
- Trapet, P.; Avoscan, L.; Klinguer, A.; Pateyron, S.; Citerne, S.; Chervin, C.; Mazurier, S.; Lemanceau, P.; Wendehenne, D.; Besson-Bard, A. The Pseudomonas Fluorescens Siderophore Pyoverdine Weakens Arabidopsis Thaliana Defense in Favor of Growth in Iron-Deficient Conditions. Plant Physiol. 2016, 171, 675–693. [Google Scholar] [CrossRef] [PubMed]
- David, S.R.; Fritsch, S.; Forster, A.; Ihiawakrim, D.; Geoffroy, V.A. Flocking Asbestos Waste, an Iron and Magnesium Source for Pseudomonas. Sci. Total Environ. 2020, 709, 135936. [Google Scholar] [CrossRef] [PubMed]
- David, S.R.; Ihiawakrim, D.; Regis, R.; Geoffroy, V.A. Efficiency of Pyoverdines in Iron Removal from Flocking Asbestos Waste: An Innovative Bacterial Bioremediation Strategy. J. Hazard. Mater. 2020, 394, 122532. [Google Scholar] [CrossRef] [PubMed]
- Moll, H.; Glorius, M.; Barkleit, A.; Rossberg, A.; Bernhard, G. The Mobilization of Actinides by Microbial Ligands Taking into Consideration the Final Storage of Nuclear Waste; Technische Informationsbibliothek: Hannover, Germany, 2009; Volume 522. [Google Scholar]
- Hazotte, A.A.; Peron, O.; Abdelouas, A.; Montavon, G.; Lebeau, T. Microbial Mobilization of Cesium from Illite: The Role of Organic Acids and Siderophores. Chem. Geol. 2016, 428, 8–14. [Google Scholar] [CrossRef]
- Péron, O.; Suzuki-Muresan, T.; Abdillahi, D.; Gaudin, P.; Abdelouas, A.; Lebeau, T. Effect of the Bacterial Pyoverdine Siderophore on the Phytoextraction of Cesium from Illite. Environ. Chem. Lett. 2019, 17, 521–526. [Google Scholar] [CrossRef]
- Nosrati, R.; Dehghani, S.; Karimi, B.; Yousefi, M.; Taghdisi, S.M.; Abnous, K.; Alibolandi, M.; Ramezani, M. Siderophore-Based Biosensors and Nanosensors; New Approach on the Development of Diagnostic Systems. Biosens. Bioelectron. 2018, 117, 1–14. [Google Scholar] [CrossRef]
- Barrero, J.M.; Morino-Bondi, M.C.; Pérez-Conde, M.C.; Cámara, C. A Biosensor for Ferric Ion. Talanta 1993, 40, 1619–1623. [Google Scholar] [CrossRef]
- Yin, K.; Zhang, W.; Chen, L. Pyoverdine Secreted by Pseudomonas aeruginosa as a Biological Recognition Element for the Fluorescent Detection of Furazolidone. Biosens. Bioelectron. 2014, 51, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Yin, K. Pyoverdine as a Biorecognition Element to Develop Biosensor for the Detection of Furazolidone. In Design of Novel Biosensors for Optical Sensing and Their Applications in Environmental Analysis; Springer: Singapore, 2020; pp. 25–35. ISBN 978-981-13-6488-4. [Google Scholar]
- Yin, K.; Wu, Y.; Wang, S.; Chen, L. A Sensitive Fluorescent Biosensor for the Detection of Copper Ion Inspired by Biological Recognition Element Pyoverdine. Sens. Actuators B Chem. 2016, 232, 257–263. [Google Scholar] [CrossRef]
- Calcott, M.J.; Owen, J.G.; Lamont, I.L.; Ackerley, D.F. Biosynthesis of Novel Pyoverdines by Domain Substitution in a Nonribosomal Peptide Synthetase of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2014, 80, 5723–5731. [Google Scholar] [CrossRef]
- Calcott, M.J.; Ackerley, D.F. Portability of the Thiolation Domain in Recombinant Pyoverdine Non-Ribosomal Peptide Synthetases. BMC Microbiol. 2015, 15, 162. [Google Scholar] [CrossRef]
- Ahmadi, M.K.; Pfeifer, B.A. Improved Heterologous Production of the Nonribosomal Peptide-Polyketide Siderophore Yersiniabactin through Metabolic Engineering and Induction Optimization. Biotechnol. Prog. 2016, 32, 1412–1417. [Google Scholar] [CrossRef]
- Qi, R.; Swayambhu, G.; Bruno, M.; Zhang, G.; Pfeifer, B.A. Consolidated Plasmid Design for Stabilized Heterologous Production of the Complex Natural Product Siderophore Yersiniabactin. Biotechnol. Prog. 2021, 37, e3103. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.J.; Kimura, N.; Sakai, A.; Ichikawa, Y.; Hanyu, T.; Otsuka, M. Cloning and Heterologous Expression of the Vibrioferrin Biosynthetic Gene Cluster from a Marine Metagenomic Library. Biosci. Biotechnol. Biochem. 2011, 75, 2283–2287. [Google Scholar] [CrossRef] [PubMed]
- Loeschcke, A.; Thies, S. Pseudomonas Putida-a Versatile Host for the Production of Natural Products. Appl. Microbiol. Biotechnol. 2015, 99, 6197–6214. [Google Scholar] [CrossRef] [PubMed]
- Baune, M.; Qi, Y.; Scholz, K.; Volmer, D.A.; Hayen, H. Structural Characterization of Pyoverdines Produced by Pseudomonas Putida KT2440 and Pseudomonas Taiwanensis VLB120. BioMetals 2017, 30, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Louden, B.C.; Haarmann, D.; Lynne, A.M. Use of Blue Agar CAS Assay for Siderophore Detection. J. Microbiol. Biol. Educ. 2011, 12, 51–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimkpa, C. Microbial Siderophores: Production, Detection and Application in Agriculture and Environment. Endocytobiosis Cell Res. 2016, 27, 7–16. [Google Scholar]
- Dane, P.R.; Pawar, S.P.; Kankariya, R.A.; Chaudhari, B.L. Pyoverdin Mediated Sunlight Induced Green Synthesis of Silver Nanoparticles. RSC Adv. 2016, 6, 8503–8510. [Google Scholar] [CrossRef]
- Cueva, A.R.; Pham, O.; Diaby, A.; Fleming, D.; Rumbaugh, K.P.; Fernandes, G.E. Pyoverdine Assay for Rapid and Early Detection of Pseudomonas aeruginosa in Burn Wounds. ACS Appl. Bio. Mater. 2020, 3, 5350–5356. [Google Scholar] [CrossRef]
- Gandouzi, I.; Tertis, M.; Cernat, A.; Bakhrouf, A.; Coros, M.; Pruneanu, S.; Cristea, C. Sensitive Detection of Pyoverdine with an Electrochemical Sensor Based on Electrochemically Generated Graphene Functionalized with Gold Nanoparticles. Bioelectrochemistry 2018, 120, 94–103. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Lee, S.; Chang, C.J. Analytical Methods for Imaging Metals in Biology: From Transition Metal Metabolism to Transition Metal Signaling. Anal. Chem. 2017, 89, 22–41. [Google Scholar] [CrossRef]
- Aschner, M.; Palinski, C.; Sperling, M.; Karst, U.; Schwerdtle, T.; Bornhorst, J. Imaging Metals in Caenorhabditis Elegans. Metallomics 2017, 9, 357–364. [Google Scholar] [CrossRef]
- Hayen, H.; Volmer, D.A. Rapid Identification of Siderophores by Combined Thin-Layer Chromatography/Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Rapid Commun. Mass Spectrom. 2005, 19, 711–720. [Google Scholar] [CrossRef]
- Boiteau, R.M.; Fitzsimmons, J.N.; Repeta, D.J.; Boyle, E.A. Detection of Iron Ligands in Seawater and Marine Cyanobacteria Cultures by High-Performance Liquid Chromatography-Inductively Coupled Plasma-Mass Spectrometry. Anal. Chem. 2013, 85, 4357–4362. [Google Scholar] [CrossRef]
- Pluháček, T.; Lemr, K.; Ghosh, D.; Milde, D.; Novák, J.; Havlíček, V. Characterization of Microbial Siderophores by Mass Spectrometry. Mass Spectrom. Rev. 2016, 35, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.-F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible Molecular Networking of Untargeted Mass Spectrometry Data Using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- Raab, A.; Feldmann, J.; Meharg, A.A. The Nature of Arsenic-Phytochelatin Complexes in Holcus Lanatus and Pteris Cretica. Plant Physiol. 2004, 134, 1113–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, E.-I.; Hung, C.S.; Parker, K.S.; Crowley, J.R.; Giblin, D.E.; Henderson, J.P. Metal Selectivity by the Virulence-Associated Yersiniabactin Metallophore System. Metallomics 2015, 7, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Petras, D.; Schmid, R.; Gauglitz, J.M.; Büttel, I.; Antelo, L.; Zhi, H.; Nuccio, S.-P.; Saak, C.C.; Malarney, K.P.; et al. Native Mass Spectrometry-Based Metabolomics Identifies Metal-Binding Compounds. Nat. Chem. 2022, 14, 100–109. [Google Scholar] [CrossRef]
- Baars, O.; Perlman, D.H. Small Molecule LC-MS/MS Fragmentation Data Analysis and Application to Siderophore Identification. In Applications from Engineering with MATLAB Concepts; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef]
- Rehm, K.; Vollenweider, V.; Kümmerli, R.; Bigler, L. A Comprehensive Method to Elucidate Pyoverdines Produced by Fluorescent Pseudomonas Spp. by UHPLC-HR-MS/MS. Anal. Bioanal. Chem. 2022, 414, 2671–2685. [Google Scholar] [CrossRef]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]



| Type of Siderophore | Structure | Organism | References |
|---|---|---|---|
| Hydroxamate Siderophores | |||
| Albomycins |  | Actinomyces suibtropicus | [8] |
| Alcaligin |  | Bordetella pertussis; Bordetella bronchiseptica | [9] |
| Bisucaberin |  | Alteromonas haloplanktis SB-112 | [10] |
| Coprogen |  | Trichoderm hypoxylon | [11] |
| Ferrichrome |  | Lactobacillus casei | [12] |
| Ferricrocin |  | Trichoderma virens | [13] |
| Danoxamine |  | Streptomyces violaceus DSM 8286 | [14] |
| Deferoxamine B |  | Streptomyces pilosus | [15] |
| Desferrioxamine E (nocardamine) |  | Streptomyces griseus | [16] |
| Fusarinine C |  | Fusarium roseum | [17] |
| Ornibactin |  | Burkholderia cepacia | [18] |
| Rhodotorulic acid |  | Rhodotorula pilimanae | [19] |
| Catecholate Siderophores | |||
| Azotochelin |  | Azotobacter vinelandi | [20] |
| Bacillibactin |  | Bacillus subtilis, Corynebacterium glutamicum | [21] |
| Enterobactin |  | Escherichia coli | [22] |
| Paenibactin |  | Paenibacillus elgii B69 | [23] |
| Protochelin |  | Azotobacter vinelandi | [24] |
| Salmochelin |  | Salmonella enterica | [25] |
| Vibriobactin |  | Vibrio cholerae | [26] |
| Carboxylate Siderophore | |||
| Achromobactin |  | Erwinia chrysanthemi | [27] |
| Rhizobactin |  | Rhizobium meliloti | [28] |
| Rhizoferrin |  | Rhizopus microsporus | [29] |
| Staphyloferrin A |  | Staphylococcus hyicus DSM20459 | [30] |
| Mixed Ligands | |||
| Aereobactin |  | E. coli | [31] |
| Amychelin |  | Amycolatopsis sp. AA4 | [32] |
| Azotobactin |  | Azotobacter vinelandii | [33] |
| Gobichelin A and B |  | Streptomyces sp. NRRL F-4415 | [34] |
| Mycobactins |  | Mycobacterium tuberculosis | [35] |
| Pseudochelin A |  | Pseudoalteromonas piscicida S2040 | [36] |
| Pyoverdine |  | Pseudomonas aeruginosa | [37] |
| Rhodobactin |  | Rhodococcus rhodochrous strain OFS | [38] |
| Yersiniabactin |  | Yersinia pestis | [39] |
| Pyoverdine Inhibitor | Inhibition Mechanism | Reference |
|---|---|---|
| LK10: 3-hydrazinylquinoxaline-2-thiol | Pyoverdine blocker | [111] |
| LK11: 1,2,3,6,7,8-hexahydro-pyrene-1,3,6,8-tetrone | Pyoverdine blocker | |
| LK12:3-amino1,4-dihydroxy-quinoxaline-2-carbonitrileN | Pyoverdine blocker | |
| LK31: (5E)-5-[(dimethylamino)methylidene]-3-(methyl-sulfanyl)-4,5,6,7-tetrahydro-2-benzothiophen-4-one | Pyoverdine blocker | |
| LK31 analog: (5E)-5-[(dimethylamino)methylidene]-3-(methylsulfanyl)- 4-oxo-4,5,6,7-tetrahydro-2-bemzothyphene-1-carbonitrile | Pyoverdine blocker | |
| PQ3: 5-oxo-3-phenyl-4-[2-(1,3-thiazol-2-yl) hydrazin-1-ylidene] pyrazole-1-carbothioamide | Pyoverdine blocker | [113] |
| PQ3 analog: (E)-3-methyl-5-oxo-4-(thiazol-2-yldiazenyl)-2,5-dihydro-1H-pyrazole-1-carbothioamide | Pyoverdine blocker | |
| Gallium | Iron-mimic | [114,115,116] |
| aPvd3 analog of PaO1 | Pyoverdine-mimic | [117] |
| 5-FC: 5-fluorocytosine | Pyoverdine biosynthesis inhibition | [102,111,118,119] |
| 5-FU: 5-fluorouracil | Pyoverdine biosynthesis inhibition | [111,118,119] |
| 5-FUR: 5-fluorouridine | Pyoverdine biosynthesis inhibition | [119,120] |
| N2- Succinyl-L-ornithine | Pyoverdine biosynthesis inhibition | [121] |
| Actinomycin X2 | Unknown | [122] |
| Actinomycin D | Unknown | |
| Eugenyl acetate | Unknown | [123] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dell’Anno, F.; Vitale, G.A.; Buonocore, C.; Vitale, L.; Palma Esposito, F.; Coppola, D.; Della Sala, G.; Tedesco, P.; de Pascale, D. Novel Insights on Pyoverdine: From Biosynthesis to Biotechnological Application. Int. J. Mol. Sci. 2022, 23, 11507. https://doi.org/10.3390/ijms231911507
Dell’Anno F, Vitale GA, Buonocore C, Vitale L, Palma Esposito F, Coppola D, Della Sala G, Tedesco P, de Pascale D. Novel Insights on Pyoverdine: From Biosynthesis to Biotechnological Application. International Journal of Molecular Sciences. 2022; 23(19):11507. https://doi.org/10.3390/ijms231911507
Chicago/Turabian StyleDell’Anno, Filippo, Giovanni Andrea Vitale, Carmine Buonocore, Laura Vitale, Fortunato Palma Esposito, Daniela Coppola, Gerardo Della Sala, Pietro Tedesco, and Donatella de Pascale. 2022. "Novel Insights on Pyoverdine: From Biosynthesis to Biotechnological Application" International Journal of Molecular Sciences 23, no. 19: 11507. https://doi.org/10.3390/ijms231911507