Does Trophectoderm Mitochondrial DNA Content Affect Embryo Developmental and Implantation Potential?
Abstract
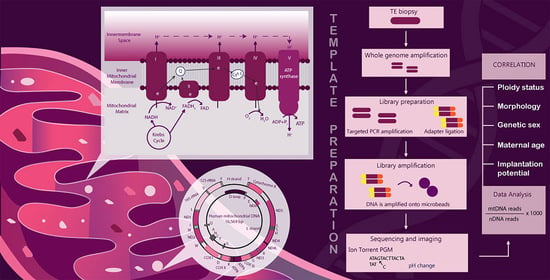
:1. Introduction
2. Results
2.1. Assocation between mtDNA Content and Embryo Chromosomal Status
2.2. Association between Maternal Age and Embryo mtDNA Levels
2.3. MtDNA Content versus Embryo Quality and Viability
2.4. The Effect of mtDNA Content on Embryo Implantation Potential
2.5. Mitochondrial Presence
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Stimulation Protocol, Oocyte Retrieval
4.3. Embryo Culture, Biopsy, and Vitrification
4.4. Whole Genome Amplification (WGA) and Next-Generation Sequencing (NGS)
4.5. Determination of Mitochondrial DNA Copy Number
4.6. Blastocyst Warming and Transfer
4.7. Cell Fluorescence Staining
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- May-Panloup, P.; Brochard, V.; Hamel, J.F.; Desquiret-Dumas, V.; Chupin, S.; Reynier, P.; Duranthon, V. Maternal Ageing Impairs Mitochondrial DNA Kinetics during Early Embryogenesis in Mice. Hum. Reprod. 2019, 34, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Boucret, L.; Bris, C.; Seegers, V.; Goudenège, D.; Desquiret-Dumas, V.; Domin-Bernhard, M.; Ferré-L’Hotellier, V.; Bouet, P.E.; Descamps, P.; Reynier, P.; et al. Deep Sequencing Shows That Oocytes Are Not Prone to Accumulate MtDNA Heteroplasmic Mutations during Ovarian Ageing. Hum. Reprod. 2017, 32, 2101–2109. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, Q.; Li, H.; Xiang, W.; Zhang, L. Cell-Free Mitochondrial DNA Increases Granulosa Cell Apoptosis and Reduces Aged Oocyte Blastocyst Development in the Mouse. Reprod. Toxicol. 2020, 98, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lin, X.; Tang, H.; Fan, Y.; Zeng, S.; Jia, L.; Li, Y.; Shi, Y.; He, S.; Wang, H.; et al. Mitochondrial DNA Mutation Exacerbates Female Reproductive Aging via Impairment of the NADH/NAD+ Redox. Aging Cell 2020, 19, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, K.F.; Machado, T.S.; Garcia, B.M.; Zangirolamo, A.F.; Macabelli, C.H.; Sugiyama, F.H.C.; Grejo, M.P.; Augusto Neto, J.D.; Tostes, K.; Ribeiro, F.K.S.; et al. Mitofusin 1 Is Required for Oocyte Growth and Communication with Follicular Somatic Cells. FASEB J. 2020, 34, 7644–7660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Bener, M.B.; Jiang, Z.; Wang, T.; Esencan, E.; Scott, R.; Horvath, T.; Seli, E. Mitofusin 2 Plays a Role in Oocyte and Follicle Development, and Is Required to Maintain Ovarian Follicular Reserve during Reproductive Aging. Aging 2019, 11, 3919–3938. [Google Scholar] [CrossRef]
- Zhang, M.; Bener, M.B.; Jiang, Z.; Wang, T.; Esencan, E.; Scott III, R.; Horvath, T.; Seli, E. Mitofusin 1 Is Required for Female Fertility and to Maintain Ovarian Follicular Reserve. Cell Death Dis. 2019, 10, 560. [Google Scholar] [CrossRef] [Green Version]
- Boucret, L.; Chao De La Barca, J.M.; Morinière, C.; Desquiret, V.; Ferré-L’Hôtellier, V.; Descamps, P.; Marcaillou, C.; Reynier, P.; Procaccio, V.; May-Panloup, P. Relationship between Diminished Ovarian Reserve and Mitochondrial Biogenesis in Cumulus Cells. Hum. Reprod. 2015, 30, 1653–1664. [Google Scholar] [CrossRef] [Green Version]
- Machado, T.S.; Macabelli, C.H.; del Collado, M.; Meirelles, F.V.; Guimarães, F.E.G.; Chiaratti, M.R. Evidence of Selection Against Damaged Mitochondria During Early Embryogenesis in the Mouse. Front. Genet. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Chatzovoulou, K.; Mayeur, A.; Gigarel, N.; Jabot-Hanin, F.; Hesters, L.; Munnich, A.; Frydman, N.; Bonnefont, J.P.; Steffann, J. Mitochondrial DNA Mutations Do Not Impact Early Human Embryonic Development. Mitochondrion 2021, 58, 59–63. [Google Scholar] [CrossRef]
- Mertens, J.; Regin, M.; de Munck, N.; de Deckersberg, E.C.; Belva, F.; Sermon, K.; Tournaye, H.; Blockeel, C.; van de Velde, H.; Spits, C. Mitochondrial DNA Variants Segregate during Human Preimplantation Development into Genetically Different Cell Lineages That Are Maintained Postnatally. Hum. Mol. Genet. 2022, 3, ddac059. [Google Scholar] [CrossRef]
- Shamsi, M.B.; Govindaraj, P.; Chawla, L.; Malhotra, N.; Singh, N.; Mittal, S.; Talwar, P.; Thangaraj, K.; Dada, R. Mitochondrial DNA Variations in Ova and Blastocyst: Implications in Assisted Reproduction. Mitochondrion 2013, 13, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, T.; Schuster, S.; Bonhoeffer, S. Cooperation and Competition in the Evolution of ATP-Producing Pathways. Science 2001, 292, 504–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giles, R.E.; Blanc, H.; Cann, H.M.; Wallace, D.C. Maternal Inheritance of Human Mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1980, 77, 6715–6719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podolak, A.; Woclawek-Potocka, I.; Lukaszuk, K. The Role of Mitochondria in Human Fertility and Early Embryo Development: What Can We Learn for Clinical Application of Assessing and Improving Mitochondrial DNA? Cells 2022, 11, 797. [Google Scholar] [CrossRef] [PubMed]
- Otten, A.B.C.; Smeets, H.J.M. Evolutionary Defined Role of the Mitochondrial DNA in Fertility, Disease and Ageing. Hum. Reprod. Update 2015, 21, 671–689. [Google Scholar] [CrossRef] [Green Version]
- Santos, T.A.; el Shourbagy, S.; St John, J.C. Mitochondrial Content Reflects Oocyte Variability and Fertilization Outcome. Fertil. Steril. 2006, 85, 584–591. [Google Scholar] [CrossRef] [Green Version]
- Barritt, J.A.; Kokot, M.; Cohen, J.; Steuerwald, N.; Brenner, C.A. Quantification of Human Ooplasmic Mitochondria. Reprod. Biomed Online 2002, 4, 243–247. [Google Scholar] [CrossRef]
- van Blerkom, J. Mitochondrial Function in the Human Oocyte and Embryo and Their Role in Developmental Competence. Mitochondrion 2011, 11, 797–813. [Google Scholar] [CrossRef]
- Liss, J.; Chromik, I.; Szczyglińska, J.; Jagiełło, M.; Łukaszuk, A.; Łukaszuk, K. Current Methods for Preimplantation Genetic Diagnosis. Ginekol. Pol. 2016, 87, 522–526. [Google Scholar] [CrossRef] [Green Version]
- Łukaszuk, K.; Kuczyńska, A.; Pukszta, S.; Kuć, P.; Kuczyński, W.; Łukaszuk, J.; Liss, J.; Wocławek-Potocka, I. First in the World Application of next Generation Sequencing in Preimplantation Genetic Diagnostics in Clinical Practice-a Case Report. Wiadomości Lek. 2016, 69, 105–108. [Google Scholar]
- Lukaszuk, K.; Jakiel, G.; Kuczynski, W.; Pukszta, S.; Liss, J.; Plociennik, L.; Lukaszuk, A.; Pastuszek, E. Next Generation Sequencing for Preimplantation Genetic Testing of Blastocysts Aneuploidies in Women of Different Ages. Ann. Agric. Environ. Med. 2015, 23, 163–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fragouli, E.; Spath, K.; Alfarawati, S.; Kaper, F.; Craig, A.; Michel, C.-E.; Kokocinski, F.; Cohen, J.; Munne, S.; Wells, D. Altered Levels of Mitochondrial DNA Are Associated with Female Age, Aneuploidy, and Provide an Independent Measure of Embryonic Implantation Potential. PLoS Genet. 2015, 11, e1005241. [Google Scholar] [CrossRef] [PubMed]
- Fragouli, E.; McCaffrey, C.; Ravichandran, K.; Spath, K.; Grifo, J.A.; Munné, S.; Wells, D. Clinical Implications of Mitochondrial DNA Quantification on Pregnancy Outcomes: A Blinded Prospective Non-Selection Study. Hum. Reprod. 2017, 32, 2340–2347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juan, A.D.; Rubio, C.; Marin, C.; Martinez, S.; Diaz-Gimeno, P.; Riboldi, M.; Al-Asmar, N.; Valbuena, D.; Simon, C. Mitochondrial DNA Content as a Viability Score in Human Euploid Embryos: Less Is Better. Fertil. Steril. 2015, 104, e311. [Google Scholar] [CrossRef]
- Du, S.; Huang, Z.; Lin, Y.; Sun, Y.; Chen, Q.; Pan, M.; Zheng, B. Mitochondrial DNA Copy Number in Human Blastocyst: A Novel Biomarker for the Prediction of Implantation Potential. J. Mol. Diagn. 2021, 23, 637–642. [Google Scholar] [CrossRef]
- Ravichandran, K.; McCaffrey, C.; Grifo, J.; Morales, A.; Perloe, M.; Munne, S.; Wells, D.; Fragouli, E. Mitochondrial DNA Quantification as a Tool for Embryo Viability Assessment: Retrospective Analysis of Data from Single Euploid Blastocyst Transfers. Hum. Reprod. 2017, 32, 1282–1292. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Li, J.; Wang, M.; Fang, Z.; Zheng, F.; Li, Z.; Jin, L. Normalized Mitochondrial DNA Copy Number Can Optimize Pregnancy Outcome Prediction in IVF. Reprod. Sci. 2021, 28, 1439–1446. [Google Scholar] [CrossRef]
- Wang, J.; Diao, Z.; Zhu, L.; Zhu, J.; Lin, F.; Jiang, W.; Fang, J.; Xu, Z.; Xing, J.; Zhou, J.; et al. Trophectoderm Mitochondrial DNA Content Associated with Embryo Quality and Day-5 Euploid Blastocyst Transfer Outcomes. DNA Cell Biol. 2021, 40, 643–651. [Google Scholar] [CrossRef]
- Ritu, G.; Veerasigamani, G.; Ashraf, M.C.; Singh, S.; Laheri, S.; Modi, D. No Association of Mitochondrial DNA Levels in Trophectodermal Cells with the Developmental Competence of the Blastocyst and Pregnancy Outcomes. bioRxiv 2019, 1–22. [Google Scholar] [CrossRef]
- Treff, N.R.; Zhan, Y.; Tao, X.; Olcha, M.; Han, M.; Rajchel, J.; Morrison, L.; Morin, S.J.; Scott, R.T.J. Levels of Trophectoderm Mitochondrial DNA Do Not Predict the Reproductive Potential of Sibling Embryos. Hum. Reprod. 2017, 32, 954–962. [Google Scholar] [CrossRef] [Green Version]
- El-Damen, A.; Elkhatib, I.; Bayram, A.; Arnanz, A.; Abdala, A.; Samir, S.; Lawrenz, B.; de Munck, N.; Fatemi, H.M. Does Blastocyst Mitochondrial DNA Content Affect Miscarriage Rate in Patients Undergoing Single Euploid Frozen Embryo Transfer? J. Assist. Reprod. Genet. 2021, 38, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Klimczak, A.M.; Pacheco, L.E.; Lewis, K.E.; Massahi, N.; Richards, J.P.; Kearns, W.G.; Saad, A.F.; Crochet, J.R. Embryonal Mitochondrial DNA: Relationship to Embryo Quality and Transfer Outcomes. J. Assist. Reprod. Genet. 2018, 35, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.; Zhang, Y.; Shu, M.; Wang, W.; Ren, L.; Chen, F.; Shao, L.; Lu, S.; Bo, S.; Ma, S.; et al. Comprehensive Chromosomal and Mitochondrial Copy Number Profiling in Human IVF Embryos. Reprod. Biomed. Online 2018, 36, 67–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podolak, A.; Liss, J.; Kiewisz, J.; Pukszta, S.; Cybulska, C.; Rychlowski, M.; Lukaszuk, A.; Jakiel, G.; Lukaszuk, K. Mitochondrial DNA Copy Number in Cleavage Stage Human Embryos—Impact on Infertility Outcome. Curr. Issues Mol. Biol. 2022, 44, 273–287. [Google Scholar] [CrossRef]
- Victor, A.R.; Brake, A.J.; Tyndall, J.C.; Griffin, D.K.; Zouves, C.G.; Barnes, F.L.; Viotti, M. Accurate Quantitation of Mitochondrial DNA Reveals Uniform Levels in Human Blastocysts Irrespective of Ploidy, Age, or Implantation Potential. Fertil. Steril. 2017, 107, 34–42. [Google Scholar] [CrossRef] [Green Version]
- Balaban, B.; Brison, D.; Calderón, G.; Catt, J.; Conaghan, J.; Cowan, L.; Ebner, T.; Gardner, D.; Hardarson, T.; Lundin, K.; et al. Istanbul Consensus Workshop on Embryo Assessment: Proceedings of an Expert Meeting. Reprod. Biomed. Online 2011, 22, 632–646. [Google Scholar] [CrossRef] [Green Version]
- Liss, J.; Pastuszek, E.; Pukszta, S.; Hoffmann, E.; Kuczynski, W.; Lukaszuk, A.; Lukaszuk, K. Effect of Next-Generation Sequencing in Preimplantation Genetic Testing on Live Birth Ratio. Reprod. Fertil. Dev. 2018, 30, 1720. [Google Scholar] [CrossRef]
- Cree, L.M.; Samuels, D.C.; de Sousa Lopes, S.C.; Rajasimha, H.K.; Wonnapinij, P.; Mann, J.R.; Dahl, H.-H.M.; Chinnery, P.F. A Reduction of Mitochondrial DNA Molecules during Embryogenesis Explains the Rapid Segregation of Genotypes. Nat. Genet. 2008, 40, 249–254. [Google Scholar] [CrossRef]
- St John, J.C.; Facucho-Oliveira, J.; Jiang, Y.; Kelly, R.; Salah, R. Mitochondrial DNA Transmission, Replication and Inheritance: A Journey from the Gamete through the Embryo and into Offspring and Embryonic Stem Cells. Hum. Reprod. Update 2010, 16, 488–509. [Google Scholar] [CrossRef] [Green Version]
- Jansen, R.P.S.; de Boer, K. The Bottleneck: Mitochondrial Imperatives in Oogenesis and Ovarian Follicular Fate. Mol. Cell. Endocrinol. 1998, 145, 81–88. [Google Scholar] [CrossRef]
- Reynier, P.; May-Panloup, P.; Chretien, M.-F.; Morgan, C.J.; Jean, M.; Savagner, F.; Barriere, P.; Malthiery, Y. Mitochondrial DNA Content Affects the Fertilizability of Human Oocytes. Mol. Hum. Reprod. 2001, 7, 425–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chappel, S. The Role of Mitochondria from Mature Oocyte to Viable Blastocyst. Obstet. Gynecol. Int. 2013, 2013, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, R.T., 3rd; Sun, L.; Zhan, Y.; Marin, D.; Tao, X.; Seli, E. Mitochondrial DNA Content Is Not Predictive of Reproductive Competence in Euploid Blastocysts. Reprod. Biomed. Online 2020, 41, 183–190. [Google Scholar] [CrossRef]
- Wu, F.S.-Y.; Weng, S.-P.; Shen, M.-S.; Ma, P.-C.; Wu, P.-K.; Lee, N.-C. Suboptimal Trophectoderm Mitochondrial DNA Level Is Associated with Delayed Blastocyst Development. J. Assist. Reprod. Genet. 2021, 38, 587–594. [Google Scholar] [CrossRef]
- De los Santos, M.J.; Diez Juan, A.; Mifsud, A.; Mercader, A.; Meseguer, M.; Rubio, C.; Pellicer, A. Variables Associated with Mitochondrial Copy Number in Human Blastocysts: What Can We Learn from Trophectoderm Biopsies? Fertil. Steril. 2018, 109, 110–117. [Google Scholar] [CrossRef]
- Arnanz, A.; de Munck, N.; Bayram, A.; El-Damen, A.; Abdalla, A.; ElKhatib, I.; Melado, L.; Lawrenz, B.; Fatemi, H.M. Blastocyst Mitochondrial DNA (MtDNA) Is Not Affected by Oocyte Vitrification: A Sibling Oocyte Study. J. Assist. Reprod. Genet. 2020, 37, 1387–1397. [Google Scholar] [CrossRef]
- Lee, Y.X.; Chen, C.H.; Lin, S.Y.; Lin, Y.H.; Tzeng, C.R. Adjusted Mitochondrial DNA Quantification in Human Embryos May Not Be Applicable as a Biomarker of Implantation Potential. J. Assist. Reprod. Genet. 2019, 36, 1855–1865. [Google Scholar] [CrossRef]
- Sunkara, S.K.; Siozos, A.; Bolton, V.N.; Khalaf, Y.; Braude, P.R.; El-Toukhy, T. The Influence of Delayed Blastocyst Formation on the Outcome of Frozen-Thawed Blastocyst Transfer: A Systematic Review and Meta-Analysis. Hum. Reprod. 2010, 25, 1906–1915. [Google Scholar] [CrossRef]
- Capalbo, A.; Rienzi, L.; Cimadomo, D.; Maggiulli, R.; Elliott, T.; Wright, G.; Nagy, Z.P.; Ubaldi, F.M. Correlation between Standard Blastocyst Morphology, Euploidy and Implantation: An Observational Study in Two Centers Involving 956 Screened Blastocysts. Hum. Reprod. 2014, 29, 1173–1181. [Google Scholar] [CrossRef] [Green Version]
- Yerushalmi, G.M.; Shavit, T.; Avraham, S.; Youngster, M.; Kedem, A.; Gat, I.; Dorofeyeva, U.S.; Mashiach, S.; Schiff, E.; Shulman, A.; et al. Day 5 Vitrified Blastocyst Transfer versus Day 6 Vitrified Blastocyst Transfer in Oocyte Donation Program. Sci. Rep. 2021, 11, 10715. [Google Scholar] [CrossRef]
- Haas, J.; Meriano, J.; Laskin, C.; Bentov, Y.; Barzilay, E.; Casper, R.F.; Cadesky, K. Clinical Pregnancy Rate Following Frozen Embryo Transfer Is Higher with Blastocysts Vitrified on Day 5 than on Day 6. J. Assist. Reprod. Genet. 2016, 33, 1553–1557. [Google Scholar] [CrossRef] [Green Version]
- Bourdon, M.; Pocate-Cheriet, K.; Finet de Bantel, A.; Grzegorczyk-Martin, V.; Amar Hoffet, A.; Arbo, E.; Poulain, M.; Santulli, P. Day 5 versus Day 6 Blastocyst Transfers: A Systematic Review and Meta-Analysis of Clinical Outcomes. Hum. Reprod. 2019, 34, 1948–1964. [Google Scholar] [CrossRef]
- Shapiro, B.S.; Richter, K.S.; Harris, D.C.; Daneshmand, S.T. A Comparison of Day 5 and Day 6 Blastocyst Transfers. Fertil. Steril. 2001, 75, 1126–1130. [Google Scholar] [CrossRef]
- Łukaszuk, K.; Pukszta, S.; Wells, D.; Cybulska, C.; Liss, J.; Płóciennik, Ł.; Kuczyński, W.; Zabielska, J. Routine Use of Next-Generation Sequencing for Preimplantation Genetic Diagnosis of Blastomeres Obtained from Embryos on Day 3 in Fresh in Vitro Fertilization Cycles. Fertil. Steril. 2015, 103, 1031–1036. [Google Scholar] [CrossRef] [PubMed]
- Kuwayama, M.; Vajta, G.; Kato, O.; Leibo, S.P. Highly Efficient Vitrification Method for Cryopreservation of Human Oocytes. Reprod. Biomed. Online 2005, 11, 300–308. [Google Scholar] [CrossRef]


| Variable | Analyzed Group | N (%) | Mean Ms ± SD | p Value |
|---|---|---|---|---|
| Analyzed Embryos | ||||
| Ploidy status | Euploid | 292 (40.8%) | 1.13 ± 1.37 | 0.02 |
| Aneuploid | 424 (59.2%) | 1.45 ± 1.78 | ||
| Genetic sex | With chr. Y | 324 (45.3%) | 1.27 ± 1.29 | 0.99 |
| Without chr. Y | 392 (54.7%) | 1.27 ± 1.18 | ||
| Maternal age | <37 years | 375 (53.4%) | 1.31 ± 1.41 | 0.43 |
| ≥37 years | 341 (47.6%) | 1.33 ± 1.29 | ||
| Embryo morphology (Istanbul criteria) | Good | 639 (89.2%) | 1.58 ± 2.44 | 0.12 |
| Poor | 77 (10.8%) | 2.19 ± 2.89 | ||
| TE biopsy day | Day 5 | 421 (58.8%) | 1.41 ± 1.66 | 0.001 |
| Day 6 | 295 (41.2%) | 1.19 ± 1.27 | ||
| Euploid Embryos | ||||
| Genetic sex | With chr. Y | 127 (43.5%) | 1.13 ± 1.47 | 0.82 |
| Without chr. Y | 165 (56.5%) | 1.23 ± 1.31 | ||
| Maternal age | <37 years | 172 (58.9%) | 1.16 ± 0.93 | 0.47 |
| ≥37 years | 120 (41.1%) | 1.08 ± 1.05 | ||
| Embryo morphology (Istanbul criteria) | Good | 264 (90.4%) | 1.38 ± 2.12 | 0.14 |
| Poor | 28 (9.6%) | 1.87 ± 2.56 | ||
| TE biopsy day | Day 5 | 187 (64%) | 1.21 ± 1.03 | 0.002 |
| Day 6 | 105 (36%) | 0.98 ± 0.86 | ||
| Transferred Embryos | ||||
| Implantation status | Implanted | 76 (36.4%) | 1.14 ± 0.88 | 0.39 |
| Non-implanted | 133 (63.6%) | 1.21 ± 1.16 | ||
| Implanted Embryos | ||||
| TE biopsy day | Day 5 | 53 (69.7%) | 1.27 ± 0.93 | 0.03 |
| Day 6 | 23 (30.3%) | 0.84 ± 0.71 | ||
| Indications for PGT-A |
|---|
| recurrent implantation failure (>=2) |
| recurrent miscarriage (>=2) |
| maternal age more than 35 |
| severe male factor |
| age of man more than 50 |
| fear of chromosomal abnormalities in the child |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lukaszuk, K.; Podolak, A. Does Trophectoderm Mitochondrial DNA Content Affect Embryo Developmental and Implantation Potential? Int. J. Mol. Sci. 2022, 23, 5976. https://doi.org/10.3390/ijms23115976
Lukaszuk K, Podolak A. Does Trophectoderm Mitochondrial DNA Content Affect Embryo Developmental and Implantation Potential? International Journal of Molecular Sciences. 2022; 23(11):5976. https://doi.org/10.3390/ijms23115976
Chicago/Turabian StyleLukaszuk, Krzysztof, and Amira Podolak. 2022. "Does Trophectoderm Mitochondrial DNA Content Affect Embryo Developmental and Implantation Potential?" International Journal of Molecular Sciences 23, no. 11: 5976. https://doi.org/10.3390/ijms23115976