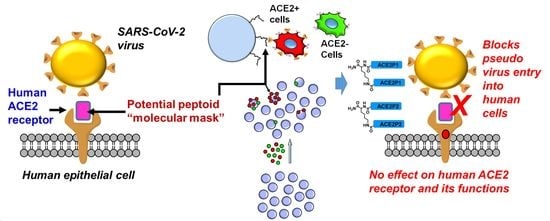
1. Introduction
Since the start of the COVID-19 pandemic, vaccine development has represented the main research and development effort to combat this deadly disease. To date, multiple vaccines have been approved by the United States Food and Drug Administration (FDA) for emergency use, and they are showing an initial success in controlling the pandemic in certain countries. Yet, the pandemic is far from over, as new strains of the SARS-CoV-2 virus with different new mutations are emerging from the continued outbreaks in various countries on different continents. The potency of current vaccines against these novel strains and the newer strains appearing in the future are not known. Reports of people who are fully vaccinated and yet are still being infected with the SARS-CoV-2 virus raise the concern over the efficacy of vaccine protection, especially with the new Delta and Lambda variants. Therefore, alternative approaches are highly warranted to find lasting solutions for both prevention and treatment of COVID-19. This is particularly important for the world to return to normal pre-pandemic life and work.
The SARS-CoV-2 virus that is causing the current global health crisis belongs to the SARS family of coronaviruses [
1,
2]. These coronaviruses (including SARS-CoV, SARS-CoV-2, MERS-CoV, and others) use their spike proteins to bind to the human ACE2 protein, which enables virus entry into human cells [
3,
4,
5,
6]. It has been known that Transmembrane Serine Protease 2 (TMPRSS2) receptor and other proteases are also involved in SARS-CoV-2 cell entry [
7]. However, TMPRSS2 acts after the virus binds to ACE2 [
3]. Without spike-ACE2 interaction, TMPRSS2 cannot allow viral entry. This is evidenced by the failure of the virus to infect ACE2 negative HeLa cells [
5], which express TMPRSS2 [
8]. In the meantime, SARS-CoV-2 can infect 293T cells expressing ACE2 but not TMPRSS2 [
3]. During SARS-CoV-2 infection, the spike protein undergoes conformational changes involving several protein domains, such as the receptor binding domain (RBD), N-terminal domain (NTD), and S1 and S2 subunits. This changes the RBD from down to up conformation and prepares the virus for binding to the ACE2 receptor and the fusion of the viral and cell membranes and the final release of the viral RNA into the cytoplasm [
9]. The mechanical stability of the RBD of SARS-CoV-2 is stiffer (greater by 50 pN) compared to SARS-CoV; thus, it can withstand Brownian and cellular forces and yet maintains close contact while priming of the spike protein by TMPRSS2 [
10]. The structure of the SARS-CoV-2 RBD-ACE2 complex has been solved using X-ray crystallography [
11]. The overall binding mode of the SARS-CoV-2 RBD to ACE2 is nearly identical to that observed in the previously determined structure of the SARS-CoV RBD-ACE2 complex. Therefore, developing molecules that can bind to ACE2 and block this interphase between viral spike protein and ACE2 can be useful as molecular blocking agents to prevent virus entry (
Figure 1a). As the family of coronaviruses has been studied for about two decades, ACE2 binding agents, including small molecules [
12,
13], peptides [
14,
15,
16,
17], and antibodies [
18] have been reported to block virus spike-ACE2 interaction [
19]. However, no ACE2-targeted compound has been approved as an antiviral drug to be used in the clinic. This may be the result of serious concerns over the inhibition of ACE2 enzymatic activity by targeting ACE2, which will interfere with the normal function of ACE2, such as blood pressure regulation [
20].
The conventional ligand or drug-lead discovery tools such as structure-based or high throughput screening (HTS) often identify molecules that bind to the active site of the targeted protein. These methods are not successful in ACE2-targeted drug discovery to block viral infection due to two reasons. First, the drug should preferably bind to the interphase where the virus spike protein binds, which is now known to be away from the ACE2 enzymatic active site [
21]. Second, the drug should not affect the physiological functions of ACE2; otherwise, the drug will have serious side effects (such as affecting blood pressure regulation). Almost all ACE2-targeted small molecules reported thus far seem to bind to the ACE2 active site [
12,
13], which may cause major health problems and disqualify them to be used as molecular masks. Therefore, novel approaches are needed to find synthetic molecules that bind to the ACE2 on the virus spike protein binding surface and are capable of blocking the spike protein-ACE2 interaction yet having no effect on ACE2 enzymatic activity. Recent computational studies have reported certain peptides binding to this interphase that can block the virus spike protein-ACE2 interaction [
22,
23,
24]. While certain peptides [
14,
15,
16,
17] and antibodies [
18,
19] reported seeming to bind to this interphase area, none of them has been developed into actual drugs thus far, indicating that avoiding ACE2-related side effects is a daunting task.
To overcome these hurdles in ACE2-targeted drug discovery, we have applied a unique combinatorial HTS technology that we developed previously to screen for new synthetic ACE2-binding ligands. Our on-bead two-color (OBTC) cell screening technology (
Figure 1b) identifies synthetic ligands based on target binding instead of the conventional function-based screens [
25,
26,
27]. OBTC compares two cell groups that differ only by the presence or absence of the target in real time and identifies ligands that only bind to the target and not any other common cell surface molecules. Therefore, this method guarantees the high selectivity of the identified ligand for the target. In addition, this assay allows for identifying ligands binding to anywhere on the receptor surface, preferably the open areas, making it possible to identify ligands that hit the spike protein binding interphase on ACE2. Most importantly, since we expose the ACE2 receptor under its natural expression condition on a live cell (as the whole system) to a library of synthetic ligands (displayed on beads) in this OBTC assay, the potential binding ligands may prefer to bind to the tip of the ACE2 receptor, an area where the virus also selected as its binding site in the viral evolution [
28,
29]. We chose peptoids as the synthetic ligands to target ACE2 for several reasons. Peptoids closely resemble the natural peptides [
30] and are protease-resistant, serum stable, and highly tissue-permeable [
31,
32,
33,
34,
35]. Their on-bead synthesis is straightforward [
34] and each coupling step can be completed in 30 seconds microwave pulses [
36]. We and others have demonstrated that peptoids are rich sources of protein-binding ligands [
25,
26,
27,
31,
35,
37,
38,
39] and are non-immunogenic in mice [
40]. Peptoid modifications are straightforward and have moderate clearance [
41]. Therefore, we believe peptoids can serve as a drug-like, biologically amenable, and economical class of molecules [
35,
42,
43,
44,
45] that may be developed into effective therapeutics or formulations of nasal sprays or eye drops.
3. Discussion
Binding of SARS-CoV-2 spike protein S to human ACE2 receptor is the key entry point for SARS-CoV-2 virus infection. Therefore, blocking this interphase of virus spike protein S and ACE2 is an attractive strategy to combat COVID19 pandemic. This has been successfully tested by developing neutralizing antibodies [
50] and an aptamer [
51] that can occupy the receptor-binding domains (RBDs) of the SARS-CoV-2 spike protein, which effectively blocks ACE2 recognition. However, the rapid development of various mutations in the virus can reduce the effectiveness of these antibodies or any other molecular classes that may be developed to bind to virus RBDs. The other side of this critical interphase is the ACE2 receptor. However, concerns over interfering with the normal physiological functions of ACE2 hinder the interests of directly targeting ACE2 for COVID-19 prevention and treatment. Since the 2003 SARS epidemic, researchers have identified numerous ACE2 binding ligands such as small molecules, peptides, and antibodies that block SARS-CoV virus entry [
21]. Peptides derived from the sequence of SARS-CoV spike protein or ACE2 were able to block SARS-CoV spike-ACE2 interaction. These peptides showed antiviral activity against SARS-CoV infection in vitro [
14,
15,
16,
17]. However, their effects on ACE2 activity have not been reported and they have not been tested against SARS-CoV-2 virus, whose spike protein is different from SARS-CoV virus (the RBD of SARS-CoV-2 is only about 74% homologous to that of SARS-CoV) [
52]. Peptides are also unstable in serum, making them less optimal as drug candidates. Small molecules were also identified to bind to ACE2. The strengths of using small molecules to target ACE2 include better pharmacokinetics and drug-like properties. However, they inhibit the function of ACE2 and their effects in blocking viral infection have not been tested [
12,
13]. Several monoclonal antibodies targeting the SARS-CoV-2 spike protein have been reported [
53,
54,
55,
56] and several are in clinical trials. These antibodies effectively blocked viral infection in animal models. A potential limitation of monoclonal antibodies is the unknown bioavailability of passively infused IgG in tissues affected by the disease, especially the lung, which is a key target of SARS-CoV-2 infection. The antibodies are large (~150 kD in size) molecules. In contrast, our peptoid compounds are 1–4 kD in size. An ACE2 specific antibody was used as a method to show the function of ACE2 as the receptor for the virus [
6]. However, the antibody was not tested as therapy to block virus entry. Certain ACE2 targeted antibodies inhibit ACE2 enzymatic activity [
57], making them unsuitable for COVID-19 prevention.
In this study, we have identified novel ACE2-binding peptoids that block spike-ACE2 interaction and prevent SARS-CoV-2 pseudotyped virus infection without affecting ACE2 expression and its enzymatic activity. These results suggest that it is possible to safely target ACE2 to prevent and treat COVID-19 and other coronavirus infections. Our ACE2 enzyme activity data, shown in
Figure 4, rule out the possibility of our peptoids binding to the ACE2 active site, as our peptoids have no effect on ACE2 activity. There can be two possibilities for potential binding sites for our peptoids on the ACE2 surface. Our peptoids may directly bind to the virus spike protein binding site on ACE2 and may directly block the virus spike protein binding to ACE2. Conversely, our peptoids may recognize a different binding site on ACE2 that has not been validated for any important activity yet but have an antagonizing effect on subsequent conformational changes needed for virus spike protein-ACE2 complex to facilitate virus entry into the cell. Furthermore, the possibility of ACE2 receptor destabilization, degradation, or internalization can be ruled out based on our data shown in
Figure 5a,b, which indicates no decrease of ACE2 protein upon peptoid binding.
Our OBTC assay uniquely allows exposing ACE2 receptor to the peptoid library while ACE2 is in its fully natural condition on the cell surface. We believe that these peptoids that were displayed on the library resin bead surface “saw” the ACE2 receptor exactly as it was “seen” by virus spike protein during the evolution to pick its target to enter human cells. The viral spike protein might have chosen the most exposed “tip” of the ACE2 receptor protein to establish its first direct contact [
28,
29], and the same might have happened with our peptoids displayed on resin beads. This hypothesis suggests that there is a high chance that our peptoids may bind to the spike protein binding site of the ACE2 and physically block the virus-ACE2 interaction. Our data is highly significant as a recent study has shown that the binding of SARS-CoV-2 spike protein caused an increase in ACE2 enzymatic activity [
23,
58], which may be relevant to the cardiovascular symptoms associated with COVID-19. This finding indicates that ligands binding to virus spike protein binding site on the ACE2 surface can affect the ACE2 active site, despite that these two sites are spatially separated. However, our peptoids have no effects on ACE2 activity (
Figure 4) and ACE2 protein expression (
Figure 5a,b), and have no toxicity to the three human cell lines that we have tested (
Figure 5c–e).
As described in the introduction, peptoids are highly biologically amenable and are easier and economical to optimize. This allows rapid development of formulations and products such as nasal sprays or eye drops using our peptoids, as the main sites of ACE2 receptor exposure are in the nose and the eye. In addition, these peptoids can be developed into conventional drugs (considering the high serum stability and tissue permeability of peptoids) to block the virus spreading inside the body tissues. Therefore, our peptoid lead compounds represent a new strategy to develop a first-line physical defense system to block virus entry using “molecular masks” that bind to the ACE2-virus spike protein interface while having no effect on the ACE2 enzyme active site. An advantage of this approach is that there will be no concern for any viral escape from an ACE2 binding peptoid, which cannot be achieved by the neutralizing approaches against the virus spike protein.
4. Materials and Methods
4.1. Chemicals and Reagents
TentaGel MB NH₂ resin (particle size: 140–170 µm, loading capacity: 0.2–0.3 mmol/g, 520,000 beads/g) was purchased from Rapp Polymere GmbH (Tuebingen, Germany). Rink amide resin (particle size: 100–200 mesh, loading capacity: 0.3–0.6 mmol/g) was purchased from Chem-Impex International, Inc. (Wood Dale, IL, USA). All Fmoc-protected amino acids and 2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU), Hydroxybenzotriazole (HOBt), all primary amines, bromoacetic acid, N,N-diisopropylcarbodiimide (DIC), N,N-diisopropylethylamine (DIPEA), piperidine, trifluoroacetic acid (TFA), cyanogen bromide (CNBr), Triisopropylsialine (TIS), α-cyano-4-hydroxycinnamic acid, acetonitrile (ACN), hydrochloric acid (HCl), dichloromethane (DCM), and N,N-dimethylformamide (DMF), were obtained from MilliporeSigma (Burlington, MA, USA). GIBCO enzyme free cell dissociation buffer and Qtracker Cell Labeling Kits were obtained from ThermoFisher Scientific (Waltham, MA, USA). All chemical reagents and solvents from commercial sources were used without further purification. Five-ml disposable reaction columns (CEM Corporation, Matthews, NC, USA) were used as reaction vessels for solid-phase synthesis. Syntheses of peptoids under microwave conditions were performed in a 1000 W microwave oven with 10% power. All purifications were completed on a Waters HPLC system (Waters Corporation, Milford, MA, USA). Mass spectra were recorded on an Applied Biosystems Voyager DE Pro mass spectrometer using α-cyano-4-hydroxycinnamic acid as the matrix.
4.2. Library Synthesis
The basic structure of the library consists of two amino acids followed by 6-mer diversified peptoid region. TentaGel MB NH2 6 g (140–170 μm; substitution: 0.2–0.3 mmol/g resin; Rapp Polymere, Tübingen, Germany) were swelled in N,N-dimethylformamide (DMF) for 30 min at room temperature in a 5 mL reaction column (Intavis AG, Tübingen, Germany) (200 mg of resin in each column). The DMF was drained from the reaction vessels, the resin was first coupled to Fmoc-Met-OH (5.0 equivalent (equiv.)) using 5.0 equiv. 2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and 5.0 equiv. Hydroxybenzotriazole (HOBt) as coupling reagents in the presence of 10.0 equiv. of N,N-diisopropylethylamine (DIPEA) for overnight shaking. Fmoc group was removed by treating the resins with 20% piperidine in DMF twice for 10 min. After washing the resins, Fmoc-Lys (Boc)-OH was added (for 2.0 h reaction time) and Fmoc group was removed as described previously. The rest of the synthesis was achieved using the split-pool synthesis protocol. A total of 10 different amines were chosen for the library, including N-Boc-1,4-butanediamine, allylamine, isobutylamine, 2-methoxyethylamine, 3-isopropoxypropylamine, β-alanine, (R)-(+)-α-methylbenzylamine, 4-methoxybenzylamine, piperonylamine, and furfurylamine. The resins were equally distributed into 10 batches for microwave-assisted peptoid synthesis steps. Each of the reaction batches was treated with 1.0 M bromoacetic acid in anhydrous DMF (1.0 mL) and 1.5 M DIC in anhydrous DMF (1.0 mL), gently shaken for 30 s, and microwaved (1000 W) for 15 s with the power set at 10%. The beads were shaken again for 30 s and microwaved another round as described above. The reaction columns were drained and washed with DMF (2.0 mL × 10 times). Then, each of the reaction batches was treated with 1.0 mL of 2.0 M solution of one primary amine (10 different batches treated with 10 different amines) and was placed on a shaker for 2.0 h at 25 °C. The resins were washed, pooled, and divided again equally into 10 batches and subjected to an addition of the next peptoid residue. This procedure was repeated until 6-mer peptoid region was completed. At the end of synthesis, the beads were washed with dichloromethane (DCM) (2.0 mL × 3 times) and treated with 2.5 mL of 95% trifluoroacetic acid (TFA), 2.5% water, and 2.5% Triisopropylsialine (TIS) on the shaker for 2 h to remove the side chain protection and were neutralized with 10% diisopropylethylamine in DMF. The reaction vessel was drained, washed with DMF (2.0 mL × 3 times), and stored in anhydrous DMF at 4 °C.
4.3. On Bead Two Color Binding Assay for Combinatorial Library Screen
A total of 50,000 peptoid library beads were washed two times in DMEM medium containing 10% FBS (media) and then incubated in 1.0 mL (DMEM + 10% FBS) for 1.0 h in a polypropylene tube. ACE2 positive MCF-7cells and ACE2 negative MCF-7 cells were removed from culture plates with GIBCO enzyme-free cell dissociation buffer (ThermoFisher Scientific, Cat # 13151014) at 2.0 mL per plate for 20 min at 37 °C. Cells were washed and suspended in DMEM + 10% FBS media. Cells were counted and distributed in 1.5 mL microcentrifuge tubes with 1.0 × 106 cells in 1.0 mL of media. Then, the cell labeling procedure was conducted as follows: 1.0 μL each of Qtracker reagent A and B were mixed in 1.5 mL microcentrifuge tubes and incubated for 5.0 min at room temperature. Media (0.2 mL) was added to each tube and vortexed for 30 s. A measured amount of 1.0 × 106 cells were added to each tube containing the labeling solution and incubated at 37 °C for 60 min. ACE2 positive MCF-7 cells were labeled with Qtracker 655 (red color) (ThermoFisher Scientific, Cat# Q25021MP, Waltham, MA, USA) and ACE2 negative MCF-7 cells labeled with Qtracker 565 (green color) (ThermoFisher Scientific, Cat# Q25031MP, Waltham, MA, USA). Cells were washed twice and suspended in 1.0 mL of DMEM + 10% FBS media. Labeled cells were visualized with long-pass filter of the BX-53F fluorescence microscope (Olympus, Waltham, MA, USA) with a color camera. Both cell types were mixed thoroughly and pipetted up and down several times to break the clumps. A cell suspension mixture of 2.0 mL was added to the tube containing 50,000 beads and incubated at room temperature with gentle shaking for 1.0 h. During incubation, cell binding to the beads were checked periodically at about 15 min intervals to ensure not to over equilibrate, which could increase non-specific binding of cells to the beads. The beads were gently washed two times with DMEM + 10% FBS media and visualized under the fluorescent microscope using the long-pass filter.
4.4. Isolation and Preparation of Beads for Sequencing
Single bead containing fluorescently tagged red cells was identified using a fluorescent microscope under 10× objective magnification and removed manually with a 20 μL pipette with medium size pipette tips. Selected beads were washed three times with 1.0% SDS and boiled in the same solution for 10 min to strip off bound cells and proteins. Finally, the beads were washed three times with water. To cleave the compound from the bead and prepare it for MS/MS sequencing, cleaving solution was prepared; thus, 30 μL of cyanogen bromide (CNBr) (5.0 M in Acetonitrile (ACN)) was added to 1.0 mL of 0.1 N HCl. Cleaving solution (50 μL) was added to the 1.5 mL tube, which contained the single isolated bead. The tube was incubated at 25 °C for 4.0 h. The solution was evaporated using a freeze dryer (SP Scientific, Gardiner, NY, USA), and the cleaved compound was suspended in 20 μL of water. MS/MS sequencing data was obtained using AB Sciex TOF/TOF 5800 machine.
4.5. Validation of on Bead Two Color Binding Screening Results
After identifying the compound (ACE2P1 and ACE2P2) with MS/MS sequencing, they were resynthesized on TentaGel MB NH₂ beads. Three tubes of 25,000 beads with each compound (containing ACE2P1 and ACE2P2 compounds) were prepared by washing and incubating for 1.0 h in DMEM + 10% FBS. Two million cells each of ACE2 positive MCF-7 cells were stained red in color using Qtracker 655, and ACE2 negative MCF-7 cells were stained in green color using Qtracker 565. One million ACE2 positive MCF-7 cells (red cells) were suspended in 1.0 mL of DMEM + 10% FBS media and were added to another 25,000 beads-containing tube. One million ACE2 negative MCF-7 cells (green cells) were suspended in 1.0 mL of DMEM + 10% FBS media and were added to 25,000 beads-containing tube. To create a mixture of cells, 0.5 × 106 of red cells and 0.5 × 106 green cells were mixed together and suspended in 1.0 mL of DMEM + 10% FBS media and were added to the third tube containing 25,000 beads. The cells were incubated with the beads for 1.0 h at room temperature. The beads were gently washed twice with DMEM + 10% FBS media and visualized under the fluorescent microscope using the long-pass filter.
4.6. Synthesis of ACE2P1
ACE2P1 was synthesized on Rink amide resin (particle size: 100–200 mesh, loading capacity: 0.3–0.6 mmol/g)/TentaGel MB NH₂ resin. An amount of 100 mg of resin was taken in 5 mL reaction column, the resin was swelled in dimethylformamide (DMF) for 1.0 h prior to use, and the Fmoc group was deprotected by treating the resin with 2.0 mL of 20% piperidine solution in DMF twice for 10 min each. The resin was first coupled to Fmoc-Met-OH (5 equiv.) using 5.0 equiv. HBTU and 5.0 equiv. HOBt as coupling reagents in the presence of 10.0 equiv. of DIPEA overnight. Fmoc was removed with the method described above. Subsequent amino acid Fmoc-Lys(Boc)-OH was introduced using the same peptide-coupling protocol (HBTU/HOBt/DIPEA), washing 10 times with DMF between each reaction. After removing the Fmoc group as described above, six peptoid residues were then coupled using a two-step peptoid coupling procedure (acylation and amination) under a microwave-assisted synthesis protocol. For the acylation step, beads were treated with 1.0 M bromoacetic acid (1.0 mL) and 1.5 M DIC (1.0 mL) and microwaved at 10% power (2 × 15 s) with gentle shaking in between for 30 s. After washing with DMF, beads were treated with 1.0 mL of (R)-(+)-α-methylbenzylamine (2.0 M), and coupling was performed by shaking at 25 °C for 2 h. The procedure was repeated to attach the remaining five residues: allylamine, 4-methoxybenzylamine, (R)-(+)-α-methylbenzylamine, 2-methoxyethylamine, and isobutylamine, in order. At the end, beads were washed with dichloromethane (DCM) and dried under vacuum before cleavage. Beads were then treated with a cleaving cocktail of TFA/H2O/TIS (95%/2.5%/2.5%) for 2.0 h. The crude compound was then purified using HPLC (Waters Corporation, Milford, MA, USA) and analyzed by MALDI-TOF (Applied Biosystems Voyager DE Pro mass spectrometer, Waltham, MA, USA). Structures are shown in
Supplementary Materials (Figures S4 and S5).
4.7. Synthesis of ACE2P1D1
ACE2P1D1 was synthesized using a protocol similar to ACE2P1. An amount of 100 mg of Rink amide resin was taken in 5 mL reaction column, the resin was swelled in dimethylformamide (DMF) for 1.0 h prior to use, and the Fmoc group was deprotected; the resin was first coupled to Fmoc-Lys(Fmoc)-OH (5 equiv.) using 5.0 equiv. HBTU and 5.0 equiv. HOBt as coupling reagents in the presence of 10.0 equiv. of DIPEA overnight. Next, both Fmoc groups were deprotected simultaneously, the Fmoc deprotection of both amine groups produced two NH2 function groups to build two copies of ACE2P1 simultaneously to obtain a homo-dimer. The resin was coupled with Fmoc-Met-OH and Fmoc-Lys(Boc)-OH using the peptide-coupling protocol (HBTU/HOBt/DIPEA). After removing the Fmoc, six peptoid residues were then coupled using a two-step peptoid coupling procedure (acylation and amination) under a microwave assisted synthesis protocol. The sequence of peptoid residues was: (R)-(+)-α-methylbenzylamine, allylamine, 4-methoxybenzylamine, (R)-(+)-α-methylbenzylamine, 2-methoxyethylamine, and isobutylamine, in order). Structure is shown in
Supplementary Materials (Figure S6).
4.8. Synthesis of Biotin-ACE2P1D1
Synthesis of Biotin-ACE2P1D1 was performed using the similar protocol described for ACE2P1. The sequence for amino acid residues for Biotin-ACE2P1D1 were Fmoc-Cys(Trt)-OH, Fmoc-Lys(Fmoc)-OH, Fmoc-Met-OH, and Fmoc-Lys(Boc)-OH. After removing the Fmoc, six peptoid residues were then coupled using a two-step peptoid coupling procedure (acylation and amination) under a microwave-assisted synthesis protocol. The sequence of peptoid residues was: (R)-(+)-α-methylbenzylamine, allylamine, 4-methoxybenzylamine, (R)-(+)-α-methylbenzylamine, 2-methoxyethylamine, and isobutylamine, in order. The compound was cleaved from the beads by treating with TFA/H2O/TIS (95%/2.5%/2.5%) for 2.0 h. Cysteine-attached ACE2P1D1 was obtained by purifying the mixture using HPLC. Biotin-maleimide [N-Biotinoyl-N′-(6-maleimidohexanoyl)hydrazide] was added to the purified portion of Cysteine attached ACE2P1D1 in 1:1 equivalent ratio in water and the pH the solution was adjusted to 7, the mixture was allowed to stir overnight at 4 °C and the compound was purified using HPLC to obtain Biotin-ACE2P1D1. Structure is shown in
Supplementary Materials (Figure S7).
4.9. Synthesis of ACE2P2
ACE2P2 was synthesized using peptide-coupling and peptoid coupling protocol similar to ACE2P1. The sequence for amino acids residues for ACE2P2D1 were Fmoc-Met-OH followed by Fmoc-Lys(Boc)-OH. Next, six peptoid residues were then coupled using a two-step peptoid coupling procedure. The sequence of peptoid residues was: 2-methoxyethylamine, isobutylamine, isobutylamine, 3-isopropoxypropylamine, 4-methoxybenzylamine and 4-methoxybenzylamine, in order. Structures are shown in
Supplementary Materials (Figures S8 and S9).
4.10. Synthesis of ACE2P2D1
ACE2P2D1 was synthesized using peptide-coupling and peptoid coupling protocol similar to ACE2P1. The sequence for amino acids residues for ACE2P2D1 were Fmoc-Lys(Fmoc)-OH followed by Fmoc-Met-OH and Fmoc-Lys(Boc)-OH, in order. Next, six peptoid residues were then coupled using a two-step peptoid coupling protocol. The sequence of peptoid residues was: 2-methoxyethylamine, isobutylamine, isobutylamine, 3-isopropoxypropylamine, 4-methoxybenzylamine and 4-methoxybenzylamine, in order. Structure is shown in
Supplementary Materials (Figure S10).
4.11. Synthesis of Biotin-ACE2P2D1
Synthesis of Biotin-ACE2P2D1 was performed using the similar protocol described for ACE2P1. The sequence for amino acids residues for Biotin-ACE2P2D1 were Fmoc-Cys(Trt)-OH, Fmoc-Lys(Fmoc)-OH, Fmoc-Met-OH and Fmoc-Lys(Boc)-OH. After removing the Fmoc, six peptoid residues were then coupled using a two-step peptoid coupling procedure (acylation and amination) under a microwave-assisted synthesis protocol. The sequence of peptoid residues was: 2-methoxyethylamine, isobutylamine, isobutylamine, 3-isopropoxypropylamine, 4-methoxybenzylamine and 4-methoxybenzylamine, in order. The compound was cleaved from the beads by treating with TFA/H2O/TIS (95%/2.5%/2.5%) for 2.0 h. Cysteine attached ACE2P2D1 was obtained by purifying the mixture using HPLC. Next, Biotin-maleimide [N-Biotinoyl-N′-(6-maleimidohexanoyl)hydrazide] was added as described for Biotin-ACE2P1D1 to obtain Biotin-ACE2P2D1. Structure is shown in
Supplementary Materials (Figure S11).
4.12. ELISA-Like Assay
An amount of 100 µL/well of recombinant human angiotensin-converting enzyme 2 (ACE2) (His-tag) (RayBiotech, Cat# 230-30165-100, Peachtree Corners, GA, USA) protein (1 µg/mL in TBS) was added to nickel coated white 96-well plates (ThermoFisher Scientific, Cat# 15242, Waltham, MA, USA) and incubated for 1.0 h (hr) at room temperature (RT) with slow shaking. Wells were washed 3 × 200 µL of TBST, and 200 µL of blocking buffer (2.0% BSA in TBS) was added and the plate was incubated for 1.0 hrs at RT. Biotinylated ACE2P1D1/Biotinylated ACE2P2D1 was added with varying concentrations (diluted in blocking buffer) (100 µL/well) and incubated for 1.0 h with slow shaking, and then washed 3 × 200 µL with TBST and added 100 µL of Streptavidin-HRP (1:1000) (dilution prepared in blocking buffer) (BioLegend, Cat# 405210, San Diego, CA, USA) and incubated for 1.0 h at RT with slow shaking. Washed the wells with TBST 6 times (200 µL each) and 100 µL of SuperSignal ELISA Pico Chemiluminescent Substrate (1:1 mixture) (Thermo Fisher Scientific, Cat# 37070, Waltham, MA, USA) was added to each well and luminescence signal was detected at all wavelengths.
4.13. Cells and siRNA Transfection
The cell lines MCF-7, H1299, and Caco-2 were purchased from American Type Culture Collection. Cells were maintained in Dulbecco’s Modified Eagle’s Medium (Corning, Cat# 10-017-CV, Tewksbury, MA, USA), supplemented with 100 units/mL penicillin, 100 µg/mL streptomycin, and 10% fetal bovine serum (Corning, Cat# 35-011-CV, Tewksbury, MA, USA) at 37 °C with 5% CO2. For siRNA transfection, MCF-7 cells were seeded (2.5 × 106/dish) in a 10cm cell culture dish one day before transfection and treated with human ACE2 siRNA (Dharmacon, Cat # L-005755-00-0005, Lafayette, CO, USA) for 24 h with X-tremeGENE siRNA transfection reagent (Roche, Ref# 04476093001, St. Louis, MO, USA) according to the manufacturer’s instructions.
4.14. Western Blot Analysis
Cells were lysed in RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS in PBS). Complete protease inhibitor cocktail (Roche, St. Louis, MO, USA) was added to lysis buffer before use. Protein concentration was determined by Bio-Rad DC protein assay (Bio-Rad, Hercules, CA, USA). Protein samples were subjected to SDS-PAGE and transferred to nitrocellulose membrane. The membrane was blocked in 5% non-fat milk in PBST overnight and incubated with primary antibody and subsequently with appropriate horse radish peroxidase-conjugated secondary antibody. Signals of targeted proteins were detected by the Immun-Star HRP peroxide Luminol/Enhancer (Bio-Rad, Hercules, CA, USA) and recorded on ChemiDoc Touch Imaging System (Bio-Rad, Hercules, CA, USA). Anti-hACE2 (human ACE2 receptor) antibody was purchased from R&D system, Inc. (Cat# AF933, Minneapolis, MN, USA). Full western blot data are given in
Supplementary Material (Figures S14–S16).
4.15. In Vitro Pull-Down Assay
10 nM of recombinant human ACE2 protein (R&D system, Cat# 933-ZN-010, Minneapolis, MN, USA) was incubated with ACE2P1 and ACE2P2 conjugated beads in RIPA buffer at 4 °C for 2 hrs. The beads were then washed with RIPA buffer for 3 times, and the binding proteins were eluted with 1% SDS at 95 °C for 5 min. The yielded lysates were then mixed with 4× Laemmli Sample Buffer (loading buffer) (Bio-Rad, Cat# 1610747, Hercules, CA, USA). The samples were then applied onto 8% SDS-PAGE gel. The gels were consequently subjected to Western blotting analysis.
4.16. GST Protein Interaction Pull-Down Assay
Pierce GST Protein Interaction Pull-down kit (Thermo Fisher Scientific, Cat# 21516, Waltham, MA, USA) was used to perform GST pull-down assay following manufacturer’s protocol. Columns containing glutathione agarose resin were washed 5 times by centrifugation at 1250× g for 1 min. A total of 0.1 µg of GST-tagged recombinant SARS-CoV-2 spike protein (Proteintech, Cat# Ag30689, Rosemont, IL, USA) was added to the column and incubated at 4 °C for 1 h. While incubating the spike protein, 0.1 µg of recombinant ACE2 protein (R&D system, Cat# 933-ZN-010, Minneapolis, MN, USA) was incubated with each of ACE2P1, ACE2P2, ACE2P1D1, and ACE2P2D1 in different concentrations (0.1 µM, 1 µM, and 10 µM) in 400 µL washing solution at cold room for 1 h by tube rotator with a gentle motion. After incubation, the columns with recombinant spike protein were washed 3 times by centrifugation at 1250× g for 1 min. ACE2 protein alone or ACE2 preincubated with ACE2P1, ACE2P2, ACE2P1D1, and ACE2P2D1 were added into each column, and incubated at 4 °C for 2 h (total volume of 400 µL for each sample). After incubation, they were washed with 400 µL of washing solution for a total of 3 washes. An amount of 200 µL of elution buffer (10 mM Glutathione) was added into each column and incubated for 5 min at room temperature. The samples were centrifuged at 1250× g for 1 min and placed on ice. SDS-PAGE gel (8%) was prepared, and the eluted samples were mixed with 4× Laemmli Sample Buffer (loading buffer) (Bio-Rad, Cat# 1610747, Hercules, CA, USA). Prepared samples were applied onto the gel. Western blot was performed using anti-hACE2 antibody from R&D system (Cat# AF933, Minneapolis, MN, USA).
4.17. His-D614G Spike—ACE2 Protein Interaction Pull-Down Assay
Pierce His Protein Interaction Pull-down kit (Thermo Fisher Scientific, Cat# 21277, Waltham, MA, USA) was used to perform His pull-down assay following manufacturer’s protocol. Columns containing HisPur Cobalt resin were washed a total of 5 times by centrifugation at 1250× g for 1 min. An amount of 0.1 µg of His-tagged recombinant SARS-CoV-2 S1 (D614G) protein (Sino Biological US Inc., Cat# 40591-V08H3, Wayne, PA, USA) was added to the column and incubated at 4 °C for 1 h. While incubating the spike protein (D614G), 0.1 µg of recombinant ACE2 protein (R&D system Inc., Cat# 933-ZN-010, Minneapolis, MN, USA) was incubated with each ACE2P1D1 and ACE2P2D1 in different concentrations (10 µM, and 100 µM) in 400 µL washing solution at cold room for 1 h by tube rotator with a gentle motion. After incubation, the columns with recombinant spike protein were washed for 3 times by centrifugation at 1250× g for 1 min. ACE2 protein alone or ACE2 preincubated with ACE2P1D1 and ACE2P2D1 were added into each column and incubated at 4 °C for 2 h (total volume of 400 µL for each sample). After incubation, they were washed with 400 µL of washing solution for a total of 3 washes. An amount of 200 µL of elution buffer was added into each column and incubated for 5 min at room temperature. The samples were centrifuged at 1250× g for 1 min and placed on ice. SDS-PAGE gel (8%) was prepared, and the eluted samples were mixed with 4× Laemmli Sample Buffer (loading buffer) (Bio-Rad, Cat#1610747, Hercules, CA, USA) and heated for 5 min at 95 °C. Prepared samples were applied onto the gel. Western blot was performed using anti-hACE2 antibody from R&D system(Cat# AF933, Minneapolis, MN, USA).
4.18. ACE2 Enzyme Activity Assay
ACE2 enzyme activity was determined using recombinant ACE2 protein (R&D system Inc., Cat# 933-ZN-010, Minneapolis, MN, USA) alone or in the presence of ACE2P1D1 and ACE2P2D1 in different concentrations (0.1 µM, 1 µM, and 10 µM), and Mca-Y-V-A-D-A-P-K (Dnp)-OH, Fluorogenic Peptide Substrate VI from R&D System, Inc. (Cat# ES007, Minneapolis, MN, USA). The reaction buffer was prepared using 1M NaCl, 0.5 mM ZnCl2, 75 mM Tris, protease inhibitor (40 µL/mL) (Roche, Ref# 11836153001, St. Louis, MO, USA), and 10 µM captopril (Enzo Life Sciences Inc., Cat# ALX-270-212-G001, Farmingdale, NY, USA) with pH 7.5. 0.1 µg of recombinant human ACE2 protein and 10 µM of substrate were used for a total volume of 100 µL. ACE2 protein and each concentration of ACE2P1D1 and ACE2P2D1 were first incubated in 96-well black plate (Corning, Cat# 3915, Tewksbury, MA, USA) on the shaker at room temperature for 20 min in 60 µL volume of each sample. Then, the 10 µM of substrate was added in each sample (40 µL of volume) and incubated at 37 °C for total 140 min. ACE2 enzyme activity was measured every 20 min by fluorescence plate reader (BioTek Cytation 5 cell Imaging Multi-Mode Reader, Winooski, VT, USA) with excitation at 320 nm and emission at 405 nm.
4.19. Flow Cytometry Analysis
To evaluate the impact of compounds to ACE2 upon binding in cell lines, MCF-7, NCI-H1299, and Caco-2 cell cultures reaching 70% confluences were incubated with 10 μM ACE2P1D1 and ACE2P2D1 for 48 h and then harvested using trypsin. Briefly, cells were centrifuged and washed with FACS buffer and then fixed by methanol on ice. Cells were suspended with a primary antibody-staining mixture using 0.5 μg (Human/Mouse/Rat/Hamster ACE-2 Antibody, R&D Systems, Inc., Cat# AF933, Minneapolis, MN, USA) per sample. Samples were incubated at room temperature for 1 h followed by washing and secondary antibody (mouse anti-goat IgG-FITC, Santa Cruz, Cat# sc-2356, Dallas, TX, USA) staining. Samples were incubated for another hour and washed before being analyzed using Accuri™ C6 (BD Biosciences, Franklin Lakes, NJ, USA). Data analysis was conducted by FlowJo™ v10 software (v10.5.2, FlowJo. LLC, Ashland, OR, USA).
4.20. Protein Thermal Shift Assay
Thermal shift assay was performed using 5 µg of recombinant ACE2 protein from Ray Biotech Life, Inc. (Cat# 230-30165, Peachtree Corners, GA, USA) and 1 µM or 10 µM of ACE2P1D1 or ACE2P2D1 with the Protein Thermal Shift Dye Kit (Thermo Fisher Scientific, Ref# 4461146, Waltham, MA, USA) and QuantStudio 3 Real-Time PCR Systems (Applied Biosystems by Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer’s protocol. Protein Thermal Shift Software v1.3 (Applied Biosystems by Thermo Fisher Scientific, Waltham, MA, USA) was used for analyzing the data.
4.21. Synthesis and Production of SARS-CoV-2 and D614G Mutant Pseudovirus
SARS-CoV-2 spike sequence (GenBank: MN908947.3) was optimized for human expression and synthesized by GenScript (Piscataway, NJ, USA) with a HA tag at N-terminal. D614G spike coding sequence was modified with a D614G point mutation and a truncation of 19 amino acids at the end of C terminus, cloned into pCDNA 3.1 vector (Invitrogen). To produce D614G spike pseudovirus for reporter assay, the lentivector reporter pSIN-Luc (containing luciferase as the marker gene) and lentiviral vector packaging plasmid psPAX2 (containing HIV gag and pol genes) were co-transfected with D614G spike at a ratio of 4:3:1 by lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) in 10 cm dishes of HEK293T cells (ATCC, Manassas, VA, USA). The virus supernatant was harvested at 48 h after transfection and stored at -800C after removing cell debris with a 0.45 µM filter.
4.22. Pseudotyped Virus Infection/Blocking Assay
ACE-2 positive H1299 cells were pretreated with PBS or various concentrations of ACE2P1D1 or ACE2P2D1 for 1 h and then infected with a given amount of titrated pseudovirus for 24 h. After removing the medium at 72 h incubation at 37 °C, luciferase assay was performed with Bright-Glo™ Luciferase Assay System (Promega, Madison, WI, USA) and measured with a luciferase reader. Inhibitory effect (%) was calculated by this formula: (treated cell readings/non-treated cell readings) × 100.
4.23. Statistical Analyses
Data are expressed as mean ± SEM. Data were analyzed by Student’s t test and were considered statistically significant if p < 0.05. The survival rates of the two groups were analyzed using a log-rank test and were considered statistically significant if p < 0.05. p values are represented as precise p values or generally as * p < 0.05, ** p < 0.01, and *** p < 0.001.