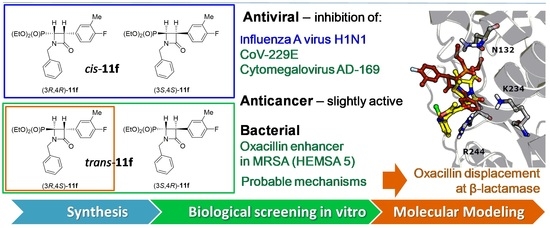
Novel N-Substituted 3-Aryl-4-(diethoxyphosphoryl)azetidin-2-ones as Antibiotic Enhancers and Antiviral Agents in Search for a Successful Treatment of Complex Infections
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Pharmacology
2.2.1. Antiviral Activity
2.2.2. Antibacterial Action
2.2.3. Antibiotics Enhancer Properties
2.2.4. Cytostatic Activity
2.3. Computer-Aided Insight into Antibiotic “Adjuvant” Action
2.3.1. Influence on β-Lactamase
2.3.2. Interactions with PBP2a
3. Materials and Methods
3.1. Chemistry
3.2. General Procedures for the Synthesis of Azetidine-2-Ones cis-10/trans-10 and cis-11/trans-11
3.2.1. General Procedure A
3.2.2. General Procedure B
3.2.3. cis-N-methyl-3-phenyl-4-(diethoxyphosphoryl)azetidin-2-one (cis-10a)
3.2.4. trans-N-methyl-3-phenyl-4-(diethoxyphosphoryl)azetidin-2-one (trans-10a)
3.2.5. cis-N-methyl-3-(2-fluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (cis-10b)
3.2.6. trans-N-methyl-3-(2-fluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (trans-10b)
3.2.7. trans-N-methyl-3-(3-fluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (trans-10c)
3.2.8. cis-N-methyl-3-(4-fluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (cis-10d)
3.2.9. trans-N-methyl-3-(4-fluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (trans-10d)
3.2.10. cis-N-methyl-3-(2,4-difluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (cis-10e)
3.2.11. trans-N-methyl-3-(2,4-difluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (trans-10e)
3.2.12. cis-N-methyl-3-(4-fluoro-3-methylphenyl)-4-(diethoxyphosphoryl)azetidin-2-one (cis-10f)
3.2.13. trans-N-methyl-3-(4-fluoro-3-methylphenyl)-4-(diethoxyphosphoryl)azetidin-2-one (trans-10f)
3.2.14. cis-N-benzyl-3-phenyl-4-(diethoxyphosphoryl)azetidin-2-one (cis-11a)
3.2.15. trans-N-benzyl-3-phenyl-4-(diethoxyphosphoryl)azetidin-2-one (trans-11a)
3.2.16. cis-N-benzyl-3-(2-fluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (cis-11b)
3.2.17. trans-N-benzyl-3-(2-fluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (trans-11b)
3.2.18. trans-N-benzyl-3-(3-fluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (trans-11c)
3.2.19. cis-N-benzyl-3-(4-fluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (cis-11d)
3.2.20. trans-N-benzyl-3-(4-fluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (trans-11d)
3.2.21. cis-N-benzyl-3-(2,4-difluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (cis-11e)
3.2.22. trans-N-benzyl-3-(2,4-difluorophenyl)-4-(diethoxyphosphoryl)azetidin-2-one (trans-11e)
3.2.23. cis-N-benzyl-3-(4-fluoro-3-methylphenyl)-4-(diethoxyphosphoryl)azetidin-2-one (cis-11f)
3.2.24. trans-N-benzyl-3-(4-fluoro-3-methylphenyl)-4-(diethoxyphosphoryl)azetidin-2-one (trans-11f)
3.3. Molecular Modelling
3.4. Antiviral Activity Assays
3.5. Cytostatic Activity against Immortalized Cell Lines
3.6. Bacterial Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of b. influenza. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Williams, J.D. β-Lactamases and β-lactamase inhibitors. Int. J. Antimicrob. Agents 1999, 12, S3–S7. [Google Scholar] [CrossRef]
- Edwards, J.R.; Betts, M.J. Carbapenems: The pinnacle of the β-lactam antibiotics or room for improvement. Int. J. Antimicrob. Chem. 2000, 45, 1–4. [Google Scholar] [CrossRef]
- Alcaide, B.; Almendros, P. Recent advances in the stereocontrolled synthesis of bi- and tricyclic-β-lactams with non-classical structure. Curr. Org. Chem. 2002, 6, 245–264. [Google Scholar] [CrossRef]
- Singh, G.S. β-Lactams in the new millennium. Part-I: Monobactams and carbapenems. Mini-Rev. Med. Chem. 2004, 4, 69–92. [Google Scholar] [CrossRef]
- Fernandes, R.; Amador, P.; Prudêncio, C. β-Lactams: Chemical structure, mode of action and mechanisms of resistance. Rev. Med. Microbiol. 2013, 24, 7–17. [Google Scholar] [CrossRef] [Green Version]
- Jarrahpour, A.; Ebrahimi, E.; Sinou, V.; Latour, C.; Brunel, J.M. Diastereoselective synthesis of potent antimalarial cis-β-lactam agents through a [2 + 2] cycloaddition of chiral imines with a chiral ketene. Eur. J. Med. Chem. 2014, 87, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Jadhav, S.; Rai, M.; Farooqui, M. Synthesis, characterization and biological evaluation of N [3-chloro-2 (aryl)-4-oxoazitidin-1-y] pyridine-4-carboxamide. Int. J. Drug Des. Discov. 2011, 2, 527–532. [Google Scholar]
- Saturnino, C.; Fusco, B.; Saturnino, P.; De Martino, G.; Rocco, F.; Lancelot, J.C. Evaluation of analgesic and anti-inflammatory activity of novel β-lactam monocyclic compounds. Biol. Pharm. Bull. 2000, 23, 654–656. [Google Scholar] [CrossRef] [Green Version]
- Zarei, M.; Mohamadzadeh, M. 3-Thiolated 2-azetidinones: Synthesis and in vitro antibacterial and antifungal activities. Tetrahedron 2011, 67, 5832–5840. [Google Scholar] [CrossRef]
- Thomas, A.B.; Nanda, R.K.; Kothapalli, L.P.; Hamane, S.C. Synthesis and biological evaluation of Schiff’s bases and 2-azetidinones of isonocotinyl hydrazone as potential antidepressant and nootropic agents. Arabian J. Chem. 2016, 9, S79–S90. [Google Scholar] [CrossRef] [Green Version]
- Dražić, T.; Sachdev, V.; Leopold, C.; Patankar, J.V.; Malnar, M.; Hećimović, S.; Levak-Frank, S.; Habuš, I.; Kratky, D. Synthesis and evaluation of novel amide amino-β-lactam derivatives as cholesterol absorption inhibitors. Bioorg. Med. Chem. 2015, 23, 2353–2359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burnett, D.A. β-Lactam cholesterol absorption inhibitors. Curr. Med. Chem. 2004, 11, 1873–1887. [Google Scholar] [CrossRef]
- Slusarchyk, W.A.; Bolton, S.A.; Hartl, K.S.; Huang, M.H.; Jacobs, G.; Meng, W.; Ogletree, M.L.; Pi, Z.; Schumacher, W.A.; Seiler, S.M.; et al. Synthesis of potent and highly selective inhibitors of human tryptase. Bioorg. Med. Chem. Lett. 2002, 12, 3235–3238. [Google Scholar] [CrossRef]
- Gupta, A.; Halve, A.K. β-Lactams: A mini review of their biological activity. Int. J. Pharm. Sci. Res. 2015, 6, 978–987. [Google Scholar] [CrossRef]
- Aoyama, Y.; Uenaka, M.; Kii, M.; Tanaka, M.; Konoike, T.; Hayasaki-Kajiwara, Y.; Nayac, N.; Nakajima, M. Design, synthesis and pharmacological evaluation of 3-benzylazetidine-2-one-based human chymase inhibitors. Bioorg. Med. Chem. 2001, 9, 3065–3075. [Google Scholar] [CrossRef]
- Guillon, C.D.; Koppel, G.A.; Brownstein, M.J.; Chaney, M.O.; Ferris, C.F.; Lu, S.; Fabio, K.M.; Miller, K.J.; Heindel, N.D.; Hunden, D.C.; et al. Azetidinones as vasopressin V1a antagonists. Bioorg. Med. Chem. 2007, 15, 2054–2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, D.-J.; Zhang, Y.-F.; Chang, A.-Q.; Li, J. β-Lactams as promising anticancer agents: Molecular hybrids, structure activity relationships and potential targets. Eur. J. Med. Chem. 2020, 201, 112510–112531. [Google Scholar] [CrossRef]
- D’hooghe, M.; Mollet, K.; De Vreese, R.; Jonckers, T.H.M.; Dams, G.; De Kimpe, N. Design, synthesis, and antiviral evaluation of purine-β-lactam and purine-aminopropanol hybrids. J. Med. Chem. 2012, 55, 5637–5641. [Google Scholar] [CrossRef]
- Smith, D.M.; Kazi, A.; Smith, L.; Long, T.E.; Heldreth, B.; Turos, E.; Dou, Q.P. A novel-lactam antibiotic activates tumor cell apoptotic program by inducing DNA damage. Mol. Pharmacol. 2002, 61, 1348–1358. [Google Scholar] [CrossRef] [Green Version]
- Chimento, A.; Sala, M.; Gomez-Monterrey, I.M.; Musella, S.; Bertamino, A.; Caruso, A.; Sinicropi, M.S.; Sirianni, R.; Puoci, F.; Parisi, O.I.; et al. Biological activity of 3-chloro-azetidin-2-one derivatives having interesting antiproliferative activity on human breast cancer cell lines. Biorg. Med. Chem. Lett. 2013, 23, 6401–6405. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Vasilevich, N.I.; Fuselier, J.A.; Hocart, S.J.; Coy, D.H. Examination of the 1,4-disubstituted azetidinone ring system as a template for combretastatin A-4 conformationally restricted analogue design. Biorg. Med. Chem. Lett. 2004, 14, 2041–2046. [Google Scholar] [CrossRef] [PubMed]
- Carr, M.; Greene, L.M.; Knox, A.J.S.; Lloyd, D.G.; Zisterer, D.M.; Meegan, M.J. Lead identification of conformationally restricted β-lactam type combretastatin analogues: Synthesis, antiproliferative activity and tubulin targeting effects. Eur. J. Med. Chem. 2010, 45, 5752–5766. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Carr, M.; Greene, L.M.; Bergin, O.; Nathwani, S.M.; McCabe, T.; Lloyd, D.G.; Zisterer, D.M.; Meegan, M.J. Synthesis and evaluation of azetidinone analogues of combretastatin A-4 as tubulin targeting agents. J. Med. Chem. 2010, 53, 8569–8584. [Google Scholar] [CrossRef] [PubMed]
- Malebari, A.M.; Fayne, D.; Nathwani, S.M.; O’Connell, F.; Noorani, S.; Twamley, B.; O’Boyle, N.M.; O’Sullivan, J.; Zisterer, D.M.; Meegan, M.J. β-Lactams with antiproliferative and antiapoptotic activity in breast and chemoresistant colon cancer cells. Eur. J. Med. Chem. 2020, 189, 112050–112075. [Google Scholar] [CrossRef]
- Gerona-Navarro, G.; Pérez de Vega, M.J.; García-López, M.T.; Andrei, G.; Snoeck, R.; Balzarini, J.; De Clercq, E.; González-Muñiz, R. Synthesis and anti-HCMV activity of 1-acyl-β-lactams and 1-acylazetidines derived from phenylalanine. Bioorg. Med. Chem. Lett. 2004, 14, 2253–2256. [Google Scholar] [CrossRef]
- Gerona-Navarro, G.; Pérez de Vega, M.J.; García-López, M.T.; Andrei, G.; Snoeck, R.; De Clercq, E.; Balzarini, J.; González-Muñiz, R. From 1-acyl-β-lactam human cytomegalovirus protease inhibitors to 1-benzyloxycarbonylazetidines with improved antiviral activity. a straightforward approach to convert covalent to noncovalent inhibitors. J. Med. Chem. 2005, 48, 2612–2621. [Google Scholar] [CrossRef]
- Dézeil, R.; Malenfant, E. Inhibition of human cytomegalovirus protease No with monocyclic β-lactams. Bioorg. Med. Chem. Lett. 1998, 8, 1437–1442. [Google Scholar] [CrossRef]
- Sperka, T.; Pitlik, J.; Bagossi, P.; Tözsér, J. Beta-lactam compounds as apparently uncompetitive inhibitors of HIV-1 protease. Bioorg. Med. Chem. Lett. 2005, 15, 3086–3090. [Google Scholar] [CrossRef] [PubMed]
- Vandekerckhove, S.; D’hooghe, M. Exploration of aziridine- and β-lactam-based hybrids as both bioactive substances and synthetic intermediates in medicinal chemistry. Bioorg. Med. Chem. 2013, 21, 3643–3647. [Google Scholar] [CrossRef] [PubMed]
- Basak, A.; Pal, R. Synthesis of β-lactam nucleoside chimera via Kinugasa reaction and evaluation of their antibacterial activity. Bioorg. Med. Chem. Lett. 2005, 15, 2015–2018. [Google Scholar] [CrossRef] [PubMed]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidta, J. ESKAPE Bacteria and Extended-Spectrum-Lactamase Producing Escherichia coli Isolated from Wastewater and Process Water from German Poultry Slaughterhouses. Appl. Environ. Microbiol. 2020, 86, e02748-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piotrowska, D.G.; Bujnowicz, A.; Wróblewski, A.E.; Głowacka, I.E. A new approach to the synthesis of 4-phosphonylated β-lactams. Synlett 2015, 26, 375–379. [Google Scholar] [CrossRef]
- Barrow, K.D.; Spotswood, T.M. Stereochemistry and P. M. R. spectra of β-lactams. Tetrahedron Lett. 1965, 6, 3325–3335. [Google Scholar] [CrossRef]
- Moriconi, E.J.; Kelly, J.F. The reaction of chlorosulfonyl isocyanate with alienes and olefins. J. Org. Chem. 1968, 33, 3036–3046. [Google Scholar] [CrossRef]
- Sterk, H.; Uray, G.; Ziegler, E. NMR-spektroskpische Untersuchungen an mehrfach substituierten β-Lactamen. Monatsh. Chem. 1972, 103, 544–550. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Dien Bard, J.; Hindler, J.A.; Gold, H.S.; Limbago, B. Rationale for eliminating Staphylococcus breakpoints for β-lactam agents other than penicillin, oxacillin or cefoxitin, and ceftaroline. Clin. Infect. Dis. 2014, 58, 1287–1296. [Google Scholar] [CrossRef]
- Piotrowska, D.G. N-Substituted C-diethoxyphosphorylated nitrones as useful synthons for the synthesis of α-aminophosphonates. Tetrahedron Lett. 2006, 47, 5363–5366. [Google Scholar] [CrossRef]
- Piotrowska, D.G.; Mediavilla, L.; Cuarental, L.; Głowacka, I.E.; Marco-Contelles, J.; Hadjipavlou-Litina, D.; López-Muñoz, F.; Oset-Gasque, M.J. Synthesis and Neuroprotective Properties of N-Substituted C-Dialkoxyphosphorylated Nitrones. ACS Omega 2019, 4, 8581–8587. [Google Scholar] [CrossRef] [Green Version]
- LigPrep, Schrödinger Release 2020-3; LLC: New York, NY, USA, 2020.
- Glide, Schrödinger Release 2020-3; LLC: New York, NY, USA, 2020.
- Protein Preparation Wizard, Schrödinger Release 2020-3; LLC: New York, NY, USA, 2020.
- Schrödinger Release 2020-3: Desmond Molecular Dynamics System; D.E. Shaw Research: New York, NY, USA; Maestro-Desmond Interoperability Tools, Schrödinger: New York, NY, USA, 2020.
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Matys, A.; Podlewska, S.; Witek, K.; Witek, J.; Bojarski, A.J.; Schabikowski, J.; Otrębska-Machaj, E.; Latacz, G.; Szymańska, E.; Kieć-Kononowicz, K.; et al. Imidazolidine-4-one derivatives in the search for novel chemosensitizers of Staphylococcus aureus MRSA: Synthesis, biological evaluation and molecular modeling studies. Eur. J. Med. Chem. 2015, 101, 313–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 9th ed.; Document M07-A9; CLSI: Wayne, PA, USA, 2012. [Google Scholar]












| Entry | Alkyne 14 (R) | 31P NMR δ [ppm] | Procedure | cis/trans Ratio 1 | Total Yield (%) 2 | Yield of cis/ trans Isomers (cis-10/trans-10) 3 | |
|---|---|---|---|---|---|---|---|
| cis-10 | trans-10 | ||||||
| a [33] |  | 19.08 | 20.52 | A | 22:78 | 76 | cis-10a–11% trans-10a–54% |
| B | 41:59 | 80 | cis-10a–21% trans-10a–40% | ||||
| b [33] |  | 18.59 | 20.06 | A | 51:49 | 84 | cis-10b–33% trans-10b–24% |
| B | 43:57 | 86 | cis-10b–19% trans-10b–42% | ||||
| c [33] |  | 18.64 | 20.10 | A | 20:80 | 74 | trans-10c–65% |
| B | 36:64 | 88 | cis-10c–7% trans-10c–55% | ||||
| d |  | 18.93 | 20.31 | A | 59:41 | 64 | cis-10d–30% trans-10d–14% |
| B | 30:70 | 65 | cis-10d–17% trans-10d–31% | ||||
| e [33] |  | 18.93 | 20.23 | A | 48:52 | 92 | cis-10e–4% trans-10e–16% |
| B | 34:66 | 60 | cis-10e–23% | ||||
| f |  | 19.04 | 20.40 | A | 37:63 | 65 | cis-10f–10% trans-10f–37% |
| B | 32:68 | 65 | cis-10f–17% trans-10f–31% | ||||
| Entry | Alkyne 14 (R) | 31P NMR δ [ppm] | Procedure | cis/trans Ratio 1 | Total Yield (%) 2 | Yield of cis/trans Isomers (cis-11/trans-11) 3 | |
|---|---|---|---|---|---|---|---|
| cis-11 | trans-11 | ||||||
| a |  | 19.15 | 20.48 | A | 21:79 | 57 | cis-11a–8% trans-11a–24% |
| B | 26:74 | 79 | cis-11a–10% trans-11a–36% | ||||
| b |  | 18.58 | 20.05 | A | 31:69 | 78 | cis-11b–20% trans-11b–26% |
| B | 30:70 | 65 | cis-11b–18% trans-11b –33% | ||||
| c |  | 20.51 | 20.90 | A | 13:87 | 63 | trans-11c–44% |
| B | 28:72 | 82 | trans-11c –56% | ||||
| d |  | 18.95 | 20.26 | A | 41:59 | 66 | cis-11d–10% trans-11d–20% |
| B | 25:75 | 54 | cis-11d–5% trans-11d–37% | ||||
| e |  | 18.41 | 19.84 | A | 68:32 | 67 | cis-11e–30% trans-11e–20% |
| B | 33:67 | 61 | cis-11e –24% trans-11e–9% | ||||
| f |  | 19.08 | 20.37 | A | 35:65 | 45 | cis-11f–7% trans-11f–14% |
| B | 30:70 | 54 | cis-11f–6% trans-11f–35% | ||||
| Cpd 1 | S. aureus ATCC 25923 | MRSA HEMSA 5 | ||||
|---|---|---|---|---|---|---|
| OXA MIC [µg/mL] | OXA+ Cpd MIC [µg/mL] | Activity Gain [A] 2 | OXA MIC [µg/mL] | OXA+ Cpd MIC [µg/mL] | Activity Gain [A] 2 | |
| trans-10a | 0.5 | 0.5 | 1 | 512 | 512 | 1 |
| cis-10a | 0.5 | 1 | 0.5 | 512 | 256–512 | 1–2 |
| trans-10b | 0.5 | 1 | 0.5 | 512 | 512 | 1 |
| cis-10b | 0.5 | 1 | 0.5 | 512 | 512 | 1 |
| trans-10c | 0.5 | 1 | 0.5 | 512 | 512 | 1 |
| trans-10d | 0.5 | 1 | 0.5 | 512 | 512 | 1 |
| cis-10d | 0.5 | 1 | 0.5 | 512 | 512 | 1 |
| trans-10e | 0.5 | >1 | <0.5 | 512 | 512 | 1 |
| cis-10e | 0.5 | 0.5 | 1 | 512 | 512 | 1 |
| trans-10f | 0.5 | 1 | 0.5 | 512 | 512 | 1 |
| cis-10f | 0.5 | 1 | 0.5 | 512 | 256–512 | 1–2 |
| trans-11a | 0.5 | >1 | <0.5 | 512 | 256–512 | 1–2 |
| cis-11a | 0.5 | 1 | 0.5 | 512 | 256–512 | 1–2 |
| trans-11b | 0.5 | >1 | <0.5 | 512 | 256–512 | 1–2 |
| cis-11b | 0.5 | 0.5 | 1 | 512 | 512 | 1 |
| trans-11c | 0.5 | >1 | <0.5 | 512 | 512 | 1 |
| trans-11d | 0.5 | 1 | 0.5 | 512 | 256 | 2 |
| cis-11d | 0.5 | 0.5 | 1 | 512 | 512 | 1 |
| trans-11e | 0.5 | 1 | 0.5 | 512 | 512 | 1 |
| cis-11e | 0.5 | 1 | 0.5 | 512 | 512 | 1 |
| trans-11f | 0.5 | 1 | 0.5 | 512 | 32 | 16 |
| cis-11f | 0.5 | >1 | <0.5 | 512 | 256–512 | 1–2 |
| Cpd | IC50 (µM) | |||||
|---|---|---|---|---|---|---|
| hTERT RPE-1 | Capan-1 | Hap1 | HCT-116 | NCI-H460 | DND-41 | |
| trans-10a | >100 | 46.2 | >100 | >100 | 54.6 | >100 |
| cis-10a | >100 | 37.8 | 46.3 | >100 | >100 | >100 |
| trans-10b | >100 | 53.1 | 38.7 | 44.1 | 34.8 | >100 |
| cis-10b | >100 | 31.9 | 14.5 | >100 | 45.2 | >100 |
| trans-10c | >100 | 19.6 | 69.1 | >100 | 62.8 | >100 |
| trans-10d | >100 | 45.7 | 58.1 | >100 | 41.3 | >100 |
| cis-10d | >100 | >100 | 70.3 | 86.5 | 71.0 | >100 |
| trans-10e | >100 | 51.7 | 51.7 | >100 | 67.6 | >100 |
| cis-10e | >100 | 56.8 | >100 | >100 | 92.6 | >100 |
| trans-10f | 90.2 | 47.7 | 41.8 | >100 | 59.7 | >100 |
| cis-10f | >100 | 61.4 | 97.9 | >100 | 87.0 | >100 |
| trans-11a | >100 | 61.0 | 63.6 | >100 | 50.9 | >100 |
| cis-11a | >100 | 95.2 | >100 | 87.9 | 78.8 | >100 |
| trans-11b | >100 | 51.9 | >100 | >100 | 56.7 | >100 |
| cis-11b | >100 | 53.0 | >100 | 74.1 | 72.8 | >100 |
| trans-11c | 45.9 | 40.1 | 56.1 | 45.5 | 44.3 | >100 |
| trans-11d | 73.0 | 35.8 | 48.3 | 44.3 | 28.4 | >100 |
| cis-11d | >100 | 35.9 | 61.9 | 54.1 | 45.9 | >100 |
| trans-11e | 25.6 | 36.5 | 42.9 | 36.6 | 24.4 | >100 |
| cis-11e | >100 | 48.9 | >100 | >100 | 43.0 | >100 |
| trans-11f | 33.5 | 34.8 | 46.5 | 35.3 | 37.0 | 65.8 |
| cis-11f | >100 | 38.4 | >100 | >100 | 53.6 | >100 |
| Docetaxel | 25.0 | 0.95 | 1.19 | 0.25 | 0.89 | 1.63 |
| Etoposide | 0.23 | 0.15 | 0.04 | 1.03 | 1.35 | 0.06 |
| Stauroporine | 0.25 | 0.66 | 3.55 | 0.09 | 11.50 | 21.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Głowacka, I.E.; Grabkowska-Drużyc, M.; Andrei, G.; Schols, D.; Snoeck, R.; Witek, K.; Podlewska, S.; Handzlik, J.; Piotrowska, D.G. Novel N-Substituted 3-Aryl-4-(diethoxyphosphoryl)azetidin-2-ones as Antibiotic Enhancers and Antiviral Agents in Search for a Successful Treatment of Complex Infections. Int. J. Mol. Sci. 2021, 22, 8032. https://doi.org/10.3390/ijms22158032
Głowacka IE, Grabkowska-Drużyc M, Andrei G, Schols D, Snoeck R, Witek K, Podlewska S, Handzlik J, Piotrowska DG. Novel N-Substituted 3-Aryl-4-(diethoxyphosphoryl)azetidin-2-ones as Antibiotic Enhancers and Antiviral Agents in Search for a Successful Treatment of Complex Infections. International Journal of Molecular Sciences. 2021; 22(15):8032. https://doi.org/10.3390/ijms22158032
Chicago/Turabian StyleGłowacka, Iwona E., Magdalena Grabkowska-Drużyc, Graciela Andrei, Dominique Schols, Robert Snoeck, Karolina Witek, Sabina Podlewska, Jadwiga Handzlik, and Dorota G. Piotrowska. 2021. "Novel N-Substituted 3-Aryl-4-(diethoxyphosphoryl)azetidin-2-ones as Antibiotic Enhancers and Antiviral Agents in Search for a Successful Treatment of Complex Infections" International Journal of Molecular Sciences 22, no. 15: 8032. https://doi.org/10.3390/ijms22158032