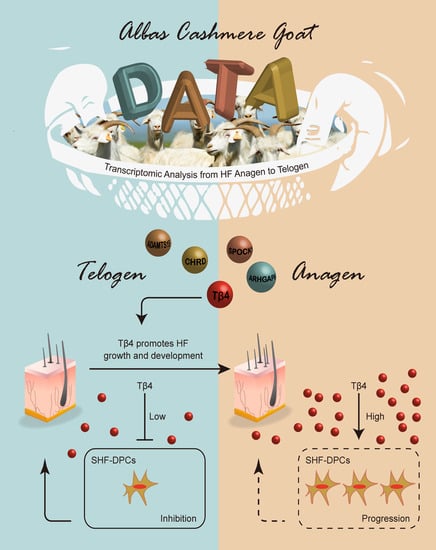
Thymosin β4 Identified by Transcriptomic Analysis from HF Anagen to Telogen Promotes Proliferation of SHF-DPCs in Albas Cashmere Goat
Abstract
:1. Introduction
2. Results
2.1. Selection of Genes from RNA-Seq from HF Anagen to Telogen of Albas Cashmere Goat
2.2. Functional Enrichment and Protein-Protein Interaction (PPI) Construction of Selected DEGs
2.3. Analysis of Relationship between Candidate Genes and Hair Follicle Growth
2.4. Identification of Tβ4 Promoting Proliferation of SHF-DPCs
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. RNA Sequencing Alignment and Transcriptomic Analysis
4.3. Function Enrichment Analyses
4.4. PPI Network Construction and Analysis of Selected DEGs
4.5. Gene Set Enrichment Analysis (GSEA)
4.6. Quantitative Real-Time PCR (qRT-PCR)
4.7. Vectors and Transfection
4.8. Western Blot Analysis
4.9. Cell Proliferation Assay
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HF | Hair follicle |
| PHF | Primary hair follicle |
| SHF | Secondary hair follicle |
| DPCs | Dermal papilla cells |
| SHF-DPCs | Dermal papilla cells of secondary hair follicle |
| DEGs | Differentially expressed genes |
References
- Liu, H.; Liu, C.; Yang, G.; Li, H.; Dai, J.; Cong, Y.; Li, X. DNA polymorphism of insulin-like growth factor-binding protein-3 gene and its association with cashmere traits in cashmere goats. Asian Australas J. Anim. Sci. 2012, 25, 1515–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.Y.; Sheng, S.D.; Hui, T.Y.; Yue, C.; Sun, J.M.; Guo, D.; Guo, S.L.; Li, B.J.; Xue, H.L.; Wang, Z.Y.; et al. An integrated analysis of cashmere fineness lncrnas in cashmere goats. Genes 2019, 10, 266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, S.; Wang, Y.; Zhou, G.; Ding, Y.; Yang, Y.; Wang, X.; Zhang, E.; Chen, Y. Synchronous profiling and analysis of mrnas and ncrnas in the dermal papilla cells from cashmere goats. BMC Genom. 2019, 20, 512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Li, Y.; Zhou, G.; Gao, Y.; Ma, S.; Chen, Y.; Song, J.; Wang, X. Whole-genome bisulfite sequencing of goat skins identifies signatures associated with hair cycling. BMC Genom. 2018, 19, 638. [Google Scholar] [CrossRef]
- Bai, W.L. Multivariate statistic analysis of morphological and ecological characters of cashmere goat populations in china. J. Anhui Agric. Sci. 2006, 34, 489. [Google Scholar]
- Xue-Feng, L.V.; Zheng, W.X. Research progress and perspective in skin follicle of cashmere goat. China Anim. Husb. Vet. Med. 2010, 37, 25–28. [Google Scholar]
- Zeder, M.A.; Hesse, B. The initial domestication of goats (capra hircus) in the zagros mountains 10,000 years ago. Science 2000, 287, 2254–2257. [Google Scholar] [CrossRef]
- Ji, X.Y.; Wang, J.X.; Liu, B.; Zheng, Z.Q.; Fu, S.Y.; Tarekegn, G.M.; Bai, X.; Bai, Y.S.; Li, H.; Zhang, W.G. Comparative transcriptome analysis reveals that a ubiquitin-mediated proteolysis pathway is important for primary and secondary hair follicle development in cashmere goats. PLoS ONE 2016, 11, e0156124. [Google Scholar] [CrossRef]
- Greco, V.; Chen, T.; Rendl, M.; Schober, M.; Pasolli, H.A.; Stokes, N.; Dela Cruz-Racelis, J.; Fuchs, E. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 2009, 4, 155–169. [Google Scholar] [CrossRef] [Green Version]
- Ge, W.; Cheng, S.F.; Dyce, P.W.; De Felici, M.; Shen, W. Skin-derived stem cells as a source of primordial germ cell- and oocyte-like cells. Cell Death Dis 2016, 7, e2471. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Jin, M.; Niu, Y.; Yan, H.; Zhou, G.; Chen, Y. Crispr/cas9-mediated vdr knockout plays an essential role in the growth of dermal papilla cells through enhanced relative genes. Peer J. 2019, 7, e7230. [Google Scholar] [CrossRef] [Green Version]
- Su, R.; Fan, Y.; Qiao, X.; Li, X.; Zhang, L.; Li, C.; Li, J. Transcriptomic analysis reveals critical genes for the hair follicle of inner mongolia cashmere goat from catagen to telogen. PLoS ONE 2018, 13, e0204404. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.Z.; Sun, H.Z.; Li, S.L.; Sang, D.; Zhang, C.H.; Jin, L.; Antonini, M.; Zhao, C.F. Effects of photoperiod on nutrient digestibility, hair follicle activity and cashmere quality in inner mongolia white cashmere goats. Asian Australas J. Anim. Sci. 2019, 32, 541–547. [Google Scholar] [CrossRef]
- Millar, S.E. Molecular mechanisms regulating hair follicle development. J. Investig. Dermatol. 2002, 118, 216–225. [Google Scholar] [CrossRef]
- Houschyar, K.S.; Borrelli, M.R.; Tapking, C.; Popp, D.; Puladi, B.; Ooms, M.; Chelliah, M.P.; Rein, S.; Pforringer, D.; Thor, D.; et al. Molecular mechanisms of hair growth and regeneration: Current understanding and novel paradigms. Dermatology 2020. [Google Scholar] [CrossRef]
- Driskell, R.R.; Clavel, C.; Rendl, M.; Watt, F.M. Hair follicle dermal papilla cells at a glance. J. Cell Sci. 2011, 124, 1179–1182. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Zhu, Y.; Liu, H.; Liu, G.; Li, F. Wnt10b promotes hair follicles growth and dermal papilla cells proliferation via wnt/beta-catenin signaling pathway in rex rabbits. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Tsai, P.-C.; Gonzalez-Celeiro, M.; Chung, O.; Boumard, B.; Perdigoto, C.N.; Ezhkova, E.; Hsu, Y.-C. Hair follicles’ transit-amplifying cells govern concurrent dermal adipocyte production through Sonic Hedgehog. Genes Dev. 2016, 30, 2325–2338. [Google Scholar] [CrossRef] [Green Version]
- Itou, T.; Ito, S.; Wakamatsu, K. Effects of aging on hair color, melanosome morphology, and melanin composition in japanese females. Int. J. Mol. Sci. 2019, 20, 3739. [Google Scholar] [CrossRef] [Green Version]
- Manning, P.L.; Edwards, N.P.; Bergmann, U.; Anne, J.; Sellers, W.I.; van Veelen, A.; Sokaras, D.; Egerton, V.M.; Alonso-Mori, R.; Ignatyev, K.; et al. Pheomelanin pigment remnants mapped in fossils of an extinct mammal. Nat. Commun. 2019, 10, 2250. [Google Scholar] [CrossRef]
- Pinheiro, F.L.; Prado, G.; Ito, S.; Simon, J.D.; Wakamatsu, K.; Anelli, L.E.; Andrade, J.A.F.; Glass, K. Chemical characterization of pterosaur melanin challenges color inferences in extinct animals. Sci. Rep. 2019, 9, 15947. [Google Scholar] [CrossRef] [Green Version]
- Galbraith, H. Fundamental hair follicle biology and fine fibre production in animals. Animal 2010, 4, 1490–1509. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.; Xu, T.; Yuan, J.; Guo, X.; Liu, D. Transcriptome sequencing reveals differences between primary and secondary hair follicle-derived dermal papilla cells of the cashmere goat (capra hircus). PLoS ONE 2013, 8, e76282. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, A.L.; Slater, F.D.; White, A. Preparation, assay, and partial purification of a thymic lymphocytopoietic factor (thymosin). Proc. Natl. Acad. Sci. USA 1966, 56, 1010–1017. [Google Scholar] [CrossRef] [Green Version]
- Grant, D.S.; Kinsella, J.L.; Kibbey, M.C.; LaFlamme, S.; Burbelo, P.D.; Goldstein, A.L.; Kleinman, H.K. Matrigel induces thymosin beta 4 gene in differentiating endothelial cells. J. Cell Sci. 1995, 108, 3685–3694. [Google Scholar]
- Goldstein, A.L.; Hannappel, E.; Kleinman, H.K. Thymosin beta4: Actin-sequestering protein moonlights to repair injured tissues. Trends Mol. Med. 2005, 11, 421–429. [Google Scholar] [CrossRef]
- Malinda, K.M.; Goldstein, A.L.; Kleinman, H.K. Thymosin beta 4 stimulates directional migration of human umbilical vein endothelial cells. FASEB J. 1997, 11, 474–481. [Google Scholar] [CrossRef]
- Philp, D.; Nguyen, M.; Scheremeta, B.; St-Surin, S.; Villa, A.M.; Orgel, A.; Kleinman, H.K.; Elkin, M. Thymosin beta4 increases hair growth by activation of hair follicle stem cells. FASEB J. 2004, 18, 385–387. [Google Scholar] [CrossRef] [Green Version]
- Mollinari, C.; Ricci-Vitiani, L.; Pieri, M.; Lucantoni, C.; Rinaldi, A.M.; Racaniello, M.; De Maria, R.; Zona, C.; Pallini, R.; Merlo, D.; et al. Downregulation of thymosin beta4 in neural progenitor grafts promotes spinal cord regeneration. J. Cell Sci. 2009, 122, 4195–4207. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Liang, H.; Hou, F.; Zhang, Z.; Nuo, M.; Guo, X.; Liu, D. Thymosin beta-4 induces mouse hair growth. PLoS ONE 2015, 10, e0130040. [Google Scholar] [CrossRef] [Green Version]
- Philp, D.; St-Surin, S.; Cha, H.J.; Moon, H.S.; Kleinman, H.K.; Elkin, M. Thymosin beta 4 induces hair growth via stem cell migration and differentiation. Ann. N. Y. Acad. Sci. 2007, 1112, 95–103. [Google Scholar] [CrossRef] [Green Version]
- Dube, K.N.; Smart, N. Thymosin beta4 and the vasculature: Multiple roles in development, repair and protection against disease. Expert Opin. Biol. Ther. 2018, 18, 131–139. [Google Scholar] [CrossRef]
- Gupta, S.; Li, L. The role of thymosin beta4 in angiotensin ii-induced cardiomyocytes growth. Expert Opin. Biol. Ther. 2018, 18, 105–110. [Google Scholar] [CrossRef]
- Lee, S.I.; Kim, D.S.; Lee, H.J.; Cha, H.J.; Kim, E.C. The role of thymosin beta 4 on odontogenic differentiation in human dental pulp cells. PLoS ONE 2013, 8, e61960. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.Y.; Hou, F.; Zhang, Z.P.; Nuo, M.T.; Liang, H.; Cang, M.; Wang, Z.G.; Wang, X.; Xu, T.; Yan, L.Y.; et al. Role of thymosin beta 4 in hair growth. Mol. Genet. Genom. 2016, 291, 1639–1646. [Google Scholar] [CrossRef]
- Li, X.; Hao, F.; Hu, X.; Wang, H.; Dai, B.; Wang, X.; Liang, H.; Cang, M.; Liu, D. Generation of tbeta4 knock-in cashmere goat using crispr/cas9. Int. J. Biol. Sci. 2019, 15, 1743–1754. [Google Scholar] [CrossRef]
- Song, S.; Yang, M.; Li, Y.; Rouzi, M.; Zhao, Q.; Pu, Y.; He, X.; Mwacharo, J.M.; Yang, N.; Ma, Y.; et al. Genome-wide discovery of lincrnas with spatiotemporal expression patterns in the skin of goat during the cashmere growth cycle. BMC Genom. 2018, 19, 495. [Google Scholar] [CrossRef]
- Ma, W.; Wang, B.; Zhang, Y.; Wang, Z.; Niu, D.; Chen, S.; Zhang, Z.; Shen, N.; Han, W.; Zhang, X.; et al. Prognostic significance of top2a in non-small cell lung cancer revealed by bioinformatic analysis. Cancer Cell Int. 2019, 19, 239. [Google Scholar] [CrossRef]
- Xu, T.; Guo, X.; Wang, H.; Du, X.; Gao, X.; Liu, D. De novo transcriptome assembly and differential gene expression profiling of three capra hircus skin types during anagen of the hair growth cycle. Int. J. Genom. 2013, 2013, 269191. [Google Scholar]
- Wang, D.G.; Xu, X.H.; Ma, H.J.; Li, C.R.; Yue, X.Z.; Gao, J.; Zhu, W.Y. Stem cell factor combined with matrix proteins regulates the attachment and migration of melanocyte precursors of human hair follicles in vitro. Biol. Pharm. Bull. 2013, 36, 1317–1325. [Google Scholar] [CrossRef] [Green Version]
- Legue, E.; Nicolas, J.F. Hair follicle renewal: Organization of stem cells in the matrix and the role of stereotyped lineages and behaviors. Development 2005, 132, 4143–4154. [Google Scholar] [CrossRef] [Green Version]
- Shirai, K.; Obara, K.; Tohgi, N.; Yamazaki, A.; Aki, R.; Hamada, Y.; Arakawa, N.; Singh, S.R.; Hoffman, R.M.; Amoh, Y. Expression of anti-aging type-xvii collagen (col17a1/bp180) in hair follicle-associated pluripotent (hap) stem cells during differentiation. Tissue Cell 2019, 59, 33–38. [Google Scholar] [CrossRef]
- Shi, B.; Ding, Q.; He, X.; Zhu, H.; Niu, Y.; Cai, B.; Cai, J.; Lei, A.; Kang, D.; Yan, H.; et al. Tbeta4-overexpression based on the piggybac transposon system in cashmere goats alters hair fiber characteristics. Transgenic Res. 2017, 26, 77–85. [Google Scholar] [CrossRef]
- Choi, N.; Sung, J.H. Udenafil induces the hair growth effect of adipose-derived stem cells. Biomol. Ther. 2019, 27, 404–413. [Google Scholar] [CrossRef]
- Perez-Meza, D.; Ziering, C.; Sforza, M.; Krishnan, G.; Ball, E.; Daniels, E. Hair follicle growth by stromal vascular fraction-enhanced adipose transplantation in baldness. Stem Cells Cloning 2017, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Suh, A.; Pham, A.; Cress, M.J.; Pincelli, T.; TerKonda, S.P.; Bruce, A.J.; Zubair, A.C.; Wolfram, J.; Shapiro, S.A. Adipose-derived cellular and cell-derived regenerative therapies in dermatology and aesthetic rejuvenation. Ageing Res. Rev. 2019, 54, 100933. [Google Scholar] [CrossRef]
- Graf-Guimaraes, C.; Mulinari-Brenner, F.; Werner, B.; Kusma, S. Platelet-rich plasma associated with hair transplants for the treatment of androgenetic alopecia showed no benefits. J. Eur. Acad. Dermatol. Venereol. 2020. [Google Scholar] [CrossRef]
- Zhu, M.; Kong, D.; Tian, R.; Pang, M.; Mo, M.; Chen, Y.; Yang, G.; Liu Cheng, H.; Lei, X.; Fang, K.; et al. Platelet sonicates activate hair follicle stem cells and mediate enhanced hair follicle regeneration. J. Cell Mol. Med. 2020, 24, 1786–1794. [Google Scholar] [CrossRef] [Green Version]
- Gentile, P.; Garcovich, S. Autologous activated platelet-rich plasma (aa-prp) and non-activated (a-prp) in hair growth: A retrospective, blinded, randomized evaluation in androgenetic alopecia. Expert Opin. Biol. Ther. 2020, 20, 327–337. [Google Scholar] [CrossRef]
- Fisher, J. Commentary on: Platelet-rich plasma and stem cells for hair growth: A review of the literature. Aesthet. Surg. J. 2020. [Google Scholar] [CrossRef]
- Karina Samudra, M.F.; Rosadi, I.; Afini, I.; Widyastuti, T.; Sobariah, S.; Remelia, M.; Puspitasari, R.L.; Rosliana, I.; Tunggadewi, T.I. Combination of the stromal vascular fraction and platelet-rich plasma accelerates the wound healing process: Pre-clinical study in a sprague-dawley rat model. Stem Cell Investig 2019, 6, 18. [Google Scholar]
- York, K.; Meah, N.; Bhoyrul, B.; Sinclair, R. Treatment review for male pattern hair-loss. Expert Opin. Pharm. 2020. [Google Scholar] [CrossRef]
- Madaan, A.; Verma, R.; Singh, A.T.; Jaggi, M. Review of hair follicle dermal papilla cells as in vitro screening model for hair growth. Int. J. Cosmet. Sci. 2018, 40, 429–450. [Google Scholar] [CrossRef] [Green Version]
- Gentile, P.; Garcovich, S. Advances in regenerative stem cell therapy in androgenic alopecia and hair loss: Wnt pathway, growth-factor, and mesenchymal stem cell signaling impact analysis on cell growth and hair follicle development. Cells 2019, 8, 466. [Google Scholar] [CrossRef] [Green Version]
- Alizée le, R.; Edith, A.; Laëtitia, M.; Elie, F.; Colin, J.; Isabelle, P.; Sylvie, B.; Brigitte, C.; Daniel, A. Extracellular vesicles from activated dermal fibroblasts stimulate hair follicle growth through dermal papilla-secreted norrin. STEM CELLS 2019, 37, 1166–1175. [Google Scholar]
- Chi, W.; Wu, E.; Morgan, B.A. Dermal papilla cell number specifies hair size, shape and cycling and its reduction causes follicular decline. Development 2013, 140, 1676–1683. [Google Scholar] [CrossRef] [Green Version]
- Rahmani, W.; Abbasi, S.; Hagner, A.; Raharjo, E.; Kumar, R.; Hotta, A.; Magness, S.; Metzger, D.; Biernaskie, J. Hair follicle dermal stem cells regenerate the dermal sheath, repopulate the dermal papilla, and modulate hair type. Dev. Cell 2014, 31, 543–558. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.I.; Yoon, H.S.; Kim, S.M.; Park, J.E.; Hyun, Y.J.; Ko, A.; Ahn, Y.S.; Koh, Y.S.; Hyun, J.W.; Yoo, E.S.; et al. Mackerel-derived fermented fish oil promotes hair growth by anagen-stimulating pathways. Int. J. Mol. Sci. 2018, 19, 2770. [Google Scholar] [CrossRef] [Green Version]
- Yu, N.; Song, Z.; Zhang, K.; Yang, X. Mad2b acts as a negative regulatory partner of tcf4 on proliferation in human dermal papilla cells. Sci. Rep. 2017, 7, 11687. [Google Scholar] [CrossRef]
- Kwack, M.H.; Seo, C.H.; Gangadaran, P.; Ahn, B.C.; Kim, M.K.; Kim, J.C.; Sung, Y.K. Exosomes derived from human dermal papilla cells promote hair growth in cultured human hair follicles and augment the hair-inductive capacity of cultured dermal papilla spheres. Exp. Dermatol. 2019, 28, 854–857. [Google Scholar] [CrossRef]
- Yan, H.; Gao, Y.; Ding, Q.; Liu, J.; Li, Y.; Jin, M.; Xu, H.; Ma, S.; Wang, X.; Zeng, W.; et al. Exosomal micro rnas derived from dermal papilla cells mediate hair follicle stem cell proliferation and differentiation. Int. J. Biol. Sci. 2019, 15, 1368–1382. [Google Scholar] [CrossRef] [Green Version]
- Pagani, A.; Aitzetmuller, M.M.; Brett, E.A.; Konig, V.; Wenny, R.; Thor, D.; Radtke, C.; Huemer, G.M.; Machens, H.G.; Duscher, D. Skin rejuvenation through hif-1alpha modulation. Plast Reconstr Surg. 2018, 141, 600e–607e. [Google Scholar] [CrossRef]
- Enshell-Seijffers, D.; Catherine, L.; Mariko, K.; Bruce, A.M. Β-catenin activity in the dermal papilla regulates morphogenesis and regeneration of hair. Dev. Cell 2010, 18, 633–642. [Google Scholar] [CrossRef] [Green Version]
- Jacobo, A.; Dasgupta, A.; Erzberger, A.; Siletti, K.; Hudspeth, A.J. Notch-mediated determination of hair-bundle polarity in mechanosensory hair cells of the zebrafish lateral line. Curr. Biol. 2019, 29, 3579–3587.e7. [Google Scholar] [CrossRef]
- Plikus, M.V.; Mayer, J.A.; de la Cruz, D.; Baker, R.E.; Maini, P.K.; Maxson, R.; Chuong, C.M. Cyclic dermal bmp signalling regulates stem cell activation during hair regeneration. Nature 2008, 451, 340–344. [Google Scholar] [CrossRef] [Green Version]
- St-Jacques, B.; Dassule, H.R.; Karavanova, I.; Botchkarev, V.A.; Li, J.; Danielian, P.S.; McMahon, J.A.; Lewis, P.M.; Paus, R.; McMahon, A.P. Sonic hedgehog signaling is essential for hair development. Curr. Biol. 1998, 8, 1058–1068. [Google Scholar] [CrossRef] [Green Version]
- Andl, T.; Reddy, S.T.; Gaddapara, T.; Millar, S.E. Wnt signals are required for the initiation of hair follicle development. Dev. Cell 2002, 2, 643–653. [Google Scholar] [CrossRef]
- Lim, C.H.; Sun, Q.; Ratti, K.; Lee, S.H.; Zheng, Y.; Takeo, M.; Lee, W.; Rabbani, P.; Plikus, M.V.; Cain, J.E.; et al. Hedgehog stimulates hair follicle neogenesis by creating inductive dermis during murine skin wound healing. Nat. Commun. 2018, 9, 4903. [Google Scholar] [CrossRef]
- Siren, J.; Valimaki, N.; Makinen, V. Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 375–388. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. Degseq: An r package for identifying differentially expressed genes from rna-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. Clusterprofiler: An r package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Walter, W.; Sanchez-Cabo, F.; Ricote, M. Goplot: An r package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef]
- Namekata, M.; Yamamoto, M.; Goitsuka, R. Nuclear localization of meis1 in dermal papilla promotes hair matrix cell proliferation in the anagen phase of hair cycle. Biochem. Biophys. Res. Commun. 2019, 519, 727–733. [Google Scholar] [CrossRef]





| Symbol | Full Name | Description |
|---|---|---|
| Tβ4 | Thymosin β 4 | Encodes an actin sequestering protein that plays a role in the regulation of actin polymerization. The protein is also involved in cell proliferation, migration, and differentiation. |
| ARHGAP6 | Rho GTPase activating protein 6 | Encodes a member of the Rho GTPase-activating proteins (rhoGAP) family of proteins that play a role in the regulation of actin polymerization at the plasma membrane during several cellular processes. |
| ADAMTS15 | ADAM metallopeptidase with thrombospondin type 1 motif, 15 | Encodes a member of the ADAMTS (ADAM metallopeptidase with thrombospondin type 1 motif) family. The encoded preproprotein is proteolytically processed to generate the mature enzyme. This gene may function as a tumor suppressor in colorectal and breast cancers. |
| CHRD | Chordin | Encodes a secreted protein that dorsalizes early vertebrate embryonic tissues by binding to ventralizing transforming growth factor-β- like (TGF-β-like) bone morphogenetic proteins and sequestering them in latent complexes. |
| SPOCK1 | SPARC (Osteonectin), cwcv- and kazal- like domains proteoglycan 1 | Encodes the protein core of a seminal plasma proteoglycan containing chondroitin- and heparan-sulfate chains. The protein’s function is unknown. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, B.; Hao, F.; Xu, T.; Zhu, B.; Ren, L.-Q.; Han, X.-Y.; Liu, D.-J. Thymosin β4 Identified by Transcriptomic Analysis from HF Anagen to Telogen Promotes Proliferation of SHF-DPCs in Albas Cashmere Goat. Int. J. Mol. Sci. 2020, 21, 2268. https://doi.org/10.3390/ijms21072268
Dai B, Hao F, Xu T, Zhu B, Ren L-Q, Han X-Y, Liu D-J. Thymosin β4 Identified by Transcriptomic Analysis from HF Anagen to Telogen Promotes Proliferation of SHF-DPCs in Albas Cashmere Goat. International Journal of Molecular Sciences. 2020; 21(7):2268. https://doi.org/10.3390/ijms21072268
Chicago/Turabian StyleDai, Bai, Fei Hao, Teng Xu, Bing Zhu, Li-Qing Ren, Xiao-Yu Han, and Dong-Jun Liu. 2020. "Thymosin β4 Identified by Transcriptomic Analysis from HF Anagen to Telogen Promotes Proliferation of SHF-DPCs in Albas Cashmere Goat" International Journal of Molecular Sciences 21, no. 7: 2268. https://doi.org/10.3390/ijms21072268