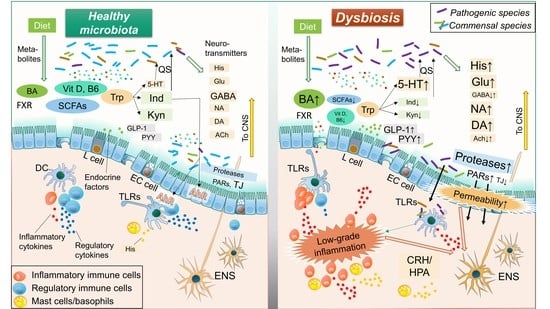
Molecular Mechanisms of Microbiota-Mediated Pathology in Irritable Bowel Syndrome
Abstract
:1. Introduction
2. Role of Intestinal Resident Microbiota in IBS Pathology
3. Mechanisms of Action: Influence of Microorganisms on Intestinal Homeostasis and IBS
3.1. Microbial Neurotransmitters
3.1.1. Histamine
3.1.2. Serotonin
3.1.3. Glutamate
3.1.4. γ-Aminobutyric Acid (GABA)
3.1.5. Others
3.2. Microbial Compounds
3.2.1. Toll-Like Receptor Ligands
3.2.2. Cytokines
3.2.3. Other Microbial Products
3.3. Microbial Metabolites
3.3.1. Tryptophan
3.3.2. Short-Chain Fatty Acids
3.3.3. Bile Acids
3.3.4. Vitamins
3.4. Microbial Endocrine Factors
3.4.1. Glucagon-Like Peptide-1 (GLP-1)
3.4.2. Peptide YY (PYY)
3.5. Microbial Enzymes: Proteases
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Enck, P.; Aziz, Q.; Barbara, G.; Farmer, A.D.; Fukudo, S.; Mayer, E.A.; Niesler, B.; Quigley, E.M.M.M.; Rajilić-Stojanović, M.; Schemann, M.; et al. Irritable bowel syndrome. Nat. Rev. Dis. Prim. 2016, 2, 16014. [Google Scholar] [CrossRef] [Green Version]
- Ford, A.C.; Lacy, B.E.; Talley, N.J. Irritable Bowel Syndrome. N. Engl. J. Med. 2017, 376, 2566–2578. [Google Scholar] [CrossRef] [Green Version]
- Sperber, A.D.; Dumitrascu, D.; Fukudo, S.; Gerson, C.; Ghoshal, U.C.; Gwee, K.A.; Hungin, A.P.S.; Kang, J.-Y.; Minhu, C.; Schmulson, M.; et al. The global prevalence of IBS in adults remains elusive due to the heterogeneity of studies: A Rome Foundation working team literature review. Gut 2017, 66, 1075–1082. [Google Scholar] [CrossRef] [Green Version]
- Schmulson, M.J.; Drossman, D.A. What Is New in Rome IV. J. Neurogastroenterol. Motil. 2017, 23, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Drossman, D.A.; Hasler, W.L. Rome IV-Functional GI Disorders: Disorders of Gut-Brain Interaction. Gastroenterology 2016, 150, 1257–1261. [Google Scholar] [CrossRef] [PubMed]
- Ford, A.C.; Harris, L.A.; Lacy, B.E.; Quigley, E.M.M.; Moayyedi, P. Systematic review with meta-analysis: The efficacy of prebiotics, probiotics, synbiotics and antibiotics in irritable bowel syndrome. Aliment. Pharmacol. Ther. 2018, 48, 1044–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camilleri, M.; Boeckxstaens, G. Dietary and pharmacological treatment of abdominal pain in IBS. Gut 2017, 66, 966–974. [Google Scholar] [CrossRef] [PubMed]
- Simrén, M.; Tack, J. New treatments and therapeutic targets for IBS and other functional bowel disorders. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 589–605. [Google Scholar] [CrossRef] [PubMed]
- Mönnikes, H. Quality of life in patients with irritable bowel syndrome. J. Clin. Gastroenterol. 2011, 45, S98–S101. [Google Scholar] [CrossRef]
- Levy, R.L.; Von Korff, M.; Whitehead, W.E.; Stang, P.; Saunders, K.; Jhingran, P.; Barghout, V.; Feld, A.D. Costs of care for irritable bowel syndrome patients in a health maintenance organization. Am. J. Gastroenterol. 2001, 96, 3122–3129. [Google Scholar] [CrossRef]
- Doshi, J.A.; Cai, Q.; Buono, J.L.; Spalding, W.M.; Sarocco, P.; Tan, H.; Stephenson, J.J.; Carson, R.T. Economic burden of irritable bowel syndrome with constipation: A retrospective analysis of health care costs in a commercially insured population. J. Manag. Care Spec. Pharm. 2014, 20, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.-Y.; Cheng, C.-W.; Tang, X.-D.; Bian, Z.-X. Impact of psychological stress on irritable bowel syndrome. World J. Gastroenterol. 2014, 20, 14126–14131. [Google Scholar] [CrossRef] [PubMed]
- Barbara, G. IBS: Biomarkers for IBS: Ready for prime time? Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Klem, F.; Wadhwa, A.; Prokop, L.J.; Sundt, W.J.; Farrugia, G.; Camilleri, M.; Singh, S.; Grover, M. Prevalence, Risk Factors, and Outcomes of Irritable Bowel Syndrome After Infectious Enteritis: A Systematic Review and Meta-analysis. Gastroenterology 2017, 152, 1042–1054.e1. [Google Scholar] [CrossRef] [Green Version]
- Vannucchi, M.G.; Evangelista, S. Experimental Models of Irritable Bowel Syndrome and the Role of the Enteric Neurotransmission. J. Clin. Med. 2018, 7, 4. [Google Scholar] [CrossRef] [Green Version]
- Qin, H.-Y.; Wu, J.C.Y.; Tong, X.-D.; Sung, J.J.Y.; Xu, H.-X.; Bian, Z.-X. Systematic review of animal models of post-infectious/post-inflammatory irritable bowel syndrome. J. Gastroenterol. 2011, 46, 164–174. [Google Scholar] [CrossRef]
- Collins, S.M. A role for the gut microbiota in IBS. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 497–505. [Google Scholar] [CrossRef]
- Gu, Y.; Zhou, G.; Qin, X.; Huang, S.; Wang, B.; Cao, H. The potential role of gut mycobiome in irritable bowel syndrome. Front. Microbiol. 2019, 10, 1894. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Godoy-Vitorino, F.; Knight, R.; Blaser, M.J. Role of the microbiome in human development. Gut 2019, 68, 1108–1114. [Google Scholar] [CrossRef]
- Sommer, F.; Anderson, J.M.; Bharti, R.; Raes, J.; Rosenstiel, P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017, 15, 630–638. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, L.; Butcher, J.; Stintzi, A.; Figeys, D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome 2019, 7, 154. [Google Scholar] [CrossRef] [PubMed]
- Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Jazwinski, S.M. The Gut Microbiota and Healthy Aging: A Mini-Review. Gerontology 2018, 64, 513–520. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Li, S.; Gan, R.-Y.; Zhou, T.; Xu, D.-P.; Li, H.-B. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef]
- Mishima, Y.; Sartor, R.B. Manipulating resident microbiota to enhance regulatory immune function to treat inflammatory bowel diseases. J. Gastroenterol. 2020, 55, 4–14. [Google Scholar] [CrossRef] [Green Version]
- Buffie, C.G.; Pamer, E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013, 13, 790–801. [Google Scholar] [CrossRef] [Green Version]
- Sorbara, M.T.; Pamer, E.G. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol. 2019, 12, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sartor, R.B.; Wu, G.D. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology 2017, 152, 327–339.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghaisas, S.; Maher, J.; Kanthasamy, A. Gut microbiome in health and disease: Linking the microbiome-gut-brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacol. Ther. 2016, 158, 52–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, M.L.; Sokol, H. The gut mycobiota: Insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 331–345. [Google Scholar] [CrossRef]
- Holtmann, G.J.; Ford, A.C.; Talley, N.J. Pathophysiology of irritable bowel syndrome. Lancet Gastroenterol. Hepatol. 2016, 1, 133–146. [Google Scholar] [CrossRef]
- Drago, L.; Valentina, C.; Fabio, P. Gut microbiota, dysbiosis and colon lavage. Dig. Liver Dis. 2019, 51, 1209–1213. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Takakura, W.; Pimentel, M. Small Intestinal Bacterial Overgrowth and Irritable Bowel Syndrome—An Update. Front. Psychiatry 2020, 11, 664. [Google Scholar] [CrossRef]
- Shah, A.; Talley, N.J.; Jones, M.; Kendall, B.J.; Koloski, N.; Walker, M.M.; Morrison, M.; Holtmann, G.J. Small Intestinal Bacterial Overgrowth in Irritable Bowel Syndrome: A Systematic Review and Meta-Analysis of Case-Control Studies. Am. J. Gastroenterol. 2020, 115, 190–201. [Google Scholar] [CrossRef]
- Kim, G.; Deepinder, F.; Morales, W.; Hwang, L.; Weitsman, S.; Chang, C.; Gunsalus, R.; Pimentel, M. Methanobrevibacter smithii is the predominant methanogen in patients with constipation-predominant IBS and methane on breath. Dig. Dis. Sci. 2012, 57, 3213–3218. [Google Scholar] [CrossRef]
- Ghoshal, U.U.C.; Shukla, R.; Srivastava, D.; Ghoshal, U.U.C. Irritable Bowel Syndrome, Particularly the Constipation-Predominant Form, Involves an Increase in Methanobrevibacter smithii, Which Is Associated with Higher Methane Production. Gut Liver 2016, 10, 932–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, T.S.B.; Raes, J.; Bork, P. The Human Gut Microbiome: From Association to Modulation. Cell 2018, 172, 1198–1215. [Google Scholar] [CrossRef] [PubMed]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, C.A.; Mu, A.; Haslam, N.; Schwartz, O.S.; Simmons, J.G. Feeling down? A systematic review of the gut microbiota in anxiety/depression and irritable bowel syndrome. J. Affect. Disord. 2020, 266, 429–446. [Google Scholar] [CrossRef] [PubMed]
- Rodiño-Janeiro, B.K.; Vicario, M.; Alonso-Cotoner, C.; Pascua-García, R.; Santos, J. A Review of Microbiota and Irritable Bowel Syndrome: Future in Therapies. Adv. Ther. 2018, 35, 289–310. [Google Scholar] [CrossRef] [Green Version]
- Duan, R.; Zhu, S.; Wang, B.; Duan, L. Alterations of Gut Microbiota in Patients With Irritable Bowel Syndrome Based on 16S rRNA-Targeted Sequencing. Clin. Transl. Gastroenterol. 2019, 10, e00012. [Google Scholar] [CrossRef]
- Wang, L.; Alammar, N.; Singh, R.; Nanavati, J.; Song, Y.; Chaudhary, R.; Mullin, G.E. Gut Microbial Dysbiosis in the Irritable Bowel Syndrome: A Systematic Review and Meta-Analysis of Case-Control Studies. J. Acad. Nutr. Diet. 2020, 120, 565–586. [Google Scholar] [CrossRef] [Green Version]
- Pittayanon, R.; Lau, J.T.; Yuan, Y.; Leontiadis, G.I.; Tse, F.; Surette, M.; Moayyedi, P. Gut Microbiota in Patients With Irritable Bowel Syndrome—A Systematic Review. Gastroenterology 2019, 157, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.-N.; Wu, H.; Chen, Y.-Z.; Chen, Y.-J.; Shen, X.-Z.; Liu, T.-T. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Dig. Liver Dis. 2017, 49, 331–337. [Google Scholar] [CrossRef]
- Crouzet, L.; Gaultier, E.; Del’Homme, C.; Cartier, C.; Delmas, E.; Dapoigny, M.; Fioramonti, J.; Bernalier-Donadille, A. The hypersensitivity to colonic distension of IBS patients can be transferred to rats through their fecal microbiota. Neurogastroenterol. Motil. 2013, 25, e272–e282. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Annamalai, C.; Rao, S.S.C. Post-Infectious Irritable Bowel Syndrome. Curr. Gastroenterol. Rep. 2017, 19, 56. [Google Scholar] [CrossRef] [PubMed]
- Hellström, P.M. Pathophysiology of the irritable bowel syndrome—Reflections of today. Best Pract. Res. Clin. Gastroenterol. 2019, 40–41, 101620. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.; Min, Y.S.; Sohn, U.D. Recent advances in pharmacological research on the management of irritable bowel syndrome. Arch. Pharm. Res. 2018, 41, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- Vandeputte, D.; Falony, G.; Vieira-Silva, S.; Tito, R.Y.; Joossens, M.; Raes, J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2016, 65, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Zeng, M.Y.; Inohara, N.; Nuñez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017, 10, 18–26. [Google Scholar] [CrossRef] [Green Version]
- Chong, P.P.; Chin, V.K.; Looi, C.Y.; Wong, W.F.; Madhavan, P.; Yong, V.C. The microbiome and irritable bowel syndrome—A review on the pathophysiology, current research and future therapy. Front. Microbiol. 2019, 10, 1136. [Google Scholar] [CrossRef] [Green Version]
- Johnsen, P.H.; Hilpüsch, F.; Valle, P.C.; Goll, R. The effect of fecal microbiota transplantation on IBS related quality of life and fatigue in moderate to severe non-constipated irritable bowel: Secondary endpoints of a double blind, randomized, placebo-controlled trial. EBioMedicine 2020, 51, 102562. [Google Scholar] [CrossRef] [Green Version]
- Wen, W.; Zhang, H.; Shen, J.; Wei, L.; Shen, S. Fecal microbiota transplantation for patients with irritable bowel syndrome: A meta-analysis protocol. Medicine (Baltim.) 2018, 97, e12661. [Google Scholar] [CrossRef]
- El-Salhy, M.; Hatlebakk, J.G.; Gilja, O.H.; Bråthen Kristoffersen, A.; Hausken, T. Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double-blind, placebo-controlled study. Gut 2020, 69, 859–867. [Google Scholar] [CrossRef] [Green Version]
- Myneedu, K.; Deoker, A.; Schmulson, M.J.; Bashashati, M. Fecal microbiota transplantation in irritable bowel syndrome: A systematic review and meta-analysis. United Eur. Gastroenterol. J. 2019, 7, 1033–1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcella, C.; Cui, B.; Kelly, C.R.; Ianiro, G.; Cammarota, G.; Zhang, F. Systematic review: The global incidence of faecal microbiota transplantation-related adverse events from 2000 to 2020. Aliment. Pharmacol. Ther. 2020. [Google Scholar] [CrossRef]
- DeFilipp, Z.; Bloom, P.P.; Torres Soto, M.; Mansour, M.K.; Sater, M.R.A.; Huntley, M.H.; Turbett, S.; Chung, R.T.; Chen, Y.-B.; Hohmann, E.L. Drug-Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N. Engl. J. Med. 2019, 381, 2043–2050. [Google Scholar] [CrossRef] [PubMed]
- Ianiro, G.; Mullish, B.H.; Kelly, C.R.; Sokol, H.; Kassam, Z.; Ng, S.C.; Fischer, M.; Allegretti, J.R.; Masucci, L.; Zhang, F.; et al. Screening of faecal microbiota transplant donors during the COVID-19 outbreak: Suggestions for urgent updates from an international expert panel. lancet. Gastroenterol. Hepatol. 2020, 5, 430–432. [Google Scholar] [CrossRef]
- Bennet, S.M.; Keshteli, A.H.; Bercik, P.; Madsen, K.L.; Reed, D.; Vanner, S.J. Application of metabolomics to the study of irritable bowel syndrome. Neurogastroenterol. Motil. 2020, 32, e13884. [Google Scholar] [CrossRef]
- Zhu, S.; Liu, S.; Li, H.; Zhang, Z.; Zhang, Q.; Chen, L.; Zhao, Y.; Chen, Y.; Gu, J.; Min, L.; et al. Identification of Gut Microbiota and Metabolites Signature in Patients With Irritable Bowel Syndrome. Front. Cell. Infect. Microbiol. 2019, 9, 346. [Google Scholar] [CrossRef] [Green Version]
- Barbara, G.; Stanghellini, V.; Brandi, G.; Cremon, C.; Di Nardo, G.; De Giorgio, R.; Corinaldesi, R. Interactions between commensal bacteria and gut sensorimotor function in health and disease. Am. J. Gastroenterol. 2005, 100, 2560–2568. [Google Scholar] [CrossRef]
- Husebye, E.; Hellström, P.M.; Sundler, F.; Chen, J.; Midtvedt, T. Influence of microbial species on small intestinal myoelectric activity and transit in germ-free rats. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 280, G368–G380. [Google Scholar] [CrossRef] [Green Version]
- Luczynski, P.; Whelan, S.O.; O’Sullivan, C.; Clarke, G.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. Adult microbiota-deficient mice have distinct dendritic morphological changes: Differential effects in the amygdala and hippocampus. Eur. J. Neurosci. 2016, 44, 2654–2666. [Google Scholar] [CrossRef] [Green Version]
- Martin, C.R.; Osadchiy, V.; Kalani, A.; Mayer, E.A. The Brain-Gut-Microbiome Axis. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 133–148. [Google Scholar] [CrossRef] [Green Version]
- Spiller, R.; Garsed, K. Postinfectious irritable bowel syndrome. Gastroenterology 2009, 136, 1979–1988. [Google Scholar] [CrossRef] [PubMed]
- Barbara, G.; Grover, M.; Bercik, P.; Corsetti, M.; Ghoshal, U.C.; Ohman, L.; Rajilić-Stojanović, M. Rome Foundation Working Team Report on Post-Infection Irritable Bowel Syndrome. Gastroenterology 2019, 156, 46–58.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shariati, A.; Fallah, F.; Pormohammad, A.; Taghipour, A.; Safari, H.; Chirani, A.S.; Sabour, S.; Alizadeh-Sani, M.; Azimi, T. The possible role of bacteria, viruses, and parasites in initiation and exacerbation of irritable bowel syndrome. J. Cell. Physiol. 2019, 234, 8550–8569. [Google Scholar] [CrossRef] [PubMed]
- Jalanka-Tuovinen, J.; Salojärvi, J.; Salonen, A.; Immonen, O.; Garsed, K.; Kelly, F.M.; Zaitoun, A.; Palva, A.; Spiller, R.C.; de Vos, W.M. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut 2014, 63, 1737–1745. [Google Scholar] [CrossRef]
- Ghoshal, U.C.; Gwee, K.A. Post-infectious IBS, tropical sprue and small intestinal bacterial overgrowth: The missing link. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 435–441. [Google Scholar] [CrossRef]
- Gupta, A.; Osadchiy, V.; Mayer, E.A. Brain-gut-microbiome interactions in obesity and food addiction. Nat. Rev. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef]
- Zhu, X.; Han, Y.; Du, J.; Liu, R.; Jin, K.; Yi, W. Microbiota-gut-brain axis and the central nervous system. Oncotarget 2017, 8, 53829–53838. [Google Scholar] [CrossRef] [Green Version]
- Barcik, W.; Wawrzyniak, M.; Akdis, C.A.; O’Mahony, L. Immune regulation by histamine and histamine-secreting bacteria. Curr. Opin. Immunol. 2017, 48, 108–113. [Google Scholar] [CrossRef]
- Wouters, M.M.; Balemans, D.; Van Wanrooy, S.; Dooley, J.; Cibert-Goton, V.; Alpizar, Y.A.; Valdez-Morales, E.E.; Nasser, Y.; Van Veldhoven, P.P.; Vanbrabant, W.; et al. Histamine Receptor H1-Mediated Sensitization of TRPV1 Mediates Visceral Hypersensitivity and Symptoms in Patients With Irritable Bowel Syndrome. Gastroenterology 2016, 150, 875–887.e9. [Google Scholar] [CrossRef] [Green Version]
- Smolinska, S.; Jutel, M.; Crameri, R.; O’Mahony, L. Histamine and gut mucosal immune regulation. Allergy 2014, 69, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Fabisiak, A.; Włodarczyk, J.; Fabisiak, N.; Storr, M.; Fichna, J. Targeting Histamine Receptors in Irritable Bowel Syndrome: A Critical Appraisal. J. Neurogastroenterol. Motil. 2017, 23, 341–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potts, R.A.; Tiffany, C.M.; Pakpour, N.; Lokken, K.L.; Tiffany, C.R.; Cheung, K.; Tsolis, R.M.; Luckhart, S. Mast cells and histamine alter intestinal permeability during malaria parasite infection. Immunobiology 2016, 221, 468–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbara, G.; Wang, B.; Stanghellini, V.; de Giorgio, R.; Cremon, C.; Di Nardo, G.; Trevisani, M.; Campi, B.; Geppetti, P.; Tonini, M.; et al. Mast cell-dependent excitation of visceral-nociceptive sensory neurons in irritable bowel syndrome. Gastroenterology 2007, 132, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Sander, L.E. Selective expression of histamine receptors H1R, H2R, and H4R, but not H3R, in the human intestinal tract. Gut 2006, 55, 498–504. [Google Scholar] [CrossRef]
- Del Colle, A.; Israelyan, N.; Gross Margolis, K. Novel aspects of enteric serotonergic signaling in health and brain-gut disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G130–G143. [Google Scholar] [CrossRef] [Green Version]
- Cremon, C.; Carini, G.; Wang, B.; Vasina, V.; Cogliandro, R.F.; De Giorgio, R.; Stanghellini, V.; Grundy, D.; Tonini, M.; De Ponti, F.; et al. Intestinal serotonin release, sensory neuron activation, and abdominal pain in irritable bowel syndrome. Am. J. Gastroenterol. 2011, 106, 1290–1298. [Google Scholar] [CrossRef]
- Knecht, L.D.; O’Connor, G.; Mittal, R.; Liu, X.Z.; Daftarian, P.; Deo, S.K.; Daunert, S. Serotonin Activates Bacterial Quorum Sensing and Enhances the Virulence of Pseudomonas aeruginosa in the Host. EBioMedicine 2016, 9, 161–169. [Google Scholar] [CrossRef] [Green Version]
- Binienda, A.; Storr, M.; Fichna, J.; Salaga, M. Efficacy and Safety of Serotonin Receptor Ligands in the Treatment of Irritable Bowel Syndrome: A Review. Curr. Drug Targets 2018, 19, 1774–1781. [Google Scholar] [CrossRef]
- McRoberts, J.A.; Coutinho, S.V.; Marvizón, J.C.; Grady, E.F.; Tognetto, M.; Sengupta, J.N.; Ennes, H.S.; Chaban, V.V.; Amadesi, S.; Creminon, C.; et al. Role of peripheral N-methyl-D-aspartate (NMDA) receptors in visceral nociception in rats. Gastroenterology 2001, 120, 1737–1748. [Google Scholar] [CrossRef]
- Filpa, V.; Moro, E.; Protasoni, M.; Crema, F.; Frigo, G.; Giaroni, C. Role of glutamatergic neurotransmission in the enteric nervous system and brain-gut axis in health and disease. Neuropharmacology 2016, 111, 14–33. [Google Scholar] [CrossRef]
- Feehily, C.; Karatzas, K.A.G. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. 2013, 114, 11–24. [Google Scholar] [CrossRef]
- Baj, A.; Moro, E.; Bistoletti, M.; Orlandi, V.; Crema, F.; Giaroni, C. Glutamatergic signaling along the microbiota-gut-brain axis. Int. J. Mol. Sci. 2019, 20, 1482. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-H.; Lin, C.-H.; Lane, H.-Y. d-glutamate and Gut Microbiota in Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 2676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Wan, J.; Rong, H.; He, F.; Wang, H.; Zhou, J.; Cai, C.; Wang, Y.; Xu, R.; Yin, Z.; et al. Alterations in Gut Glutamate Metabolism Associated with Changes in Gut Microbiota Composition in Children with Autism Spectrum Disorder. mSystems 2019, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, C.A.; Vuong, H.E.; Yano, J.M.; Liang, Q.Y.; Nusbaum, D.J.; Hsiao, E.Y. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell 2018, 173, 1728–1741.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holton, K.F.; Taren, D.L.; Thomson, C.A.; Bennett, R.M.; Jones, K.D. The effect of dietary glutamate on fibromyalgia and irritable bowel symptoms. Clin. Exp. Rheumatol. 2012, 30, 10–17. [Google Scholar]
- Qi, Q.; Chen, F.; Zhang, W.; Wang, P.; Li, Y.; Zuo, X. Colonic N-methyl-d-aspartate receptor contributes to visceral hypersensitivity in irritable bowel syndrome. J. Gastroenterol. Hepatol. 2017, 32, 828–836. [Google Scholar] [CrossRef]
- Vermeulen, M.A. Glutamate reduces experimental intestinal hyperpermeability and facilitates glutamine support of gut integrity. World J. Gastroenterol. 2011, 17, 1569. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Sun, J.; Lu, R.; Ji, Q.; Xu, J.-G. Effect of glutamate on inflammatory responses of intestine and brain after focal cerebral ischemia. World J. Gastroenterol. 2005, 11, 733–736. [Google Scholar] [CrossRef]
- de Leon, A.S.; Tadi, P. Biochemistry, Gamma Aminobutyric Acid. 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK551683/ (accessed on 15 November 2020).
- Mazzoli, R.; Pessione, E. The Neuro-endocrinological Role of Microbial Glutamate and GABA Signaling. Front. Microbiol. 2016, 7, 1934. [Google Scholar] [CrossRef] [Green Version]
- Kumar, K.; Sharma, S.; Kumar, P.; Deshmukh, R. Therapeutic potential of GABA(B) receptor ligands in drug addiction, anxiety, depression and other CNS disorders. Pharmacol. Biochem. Behav. 2013, 110, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Ahuja, V.; Paul, J. Dysregulation of GABAergic Signalling Contributes in the Pathogenesis of Diarrhea-predominant Irritable Bowel Syndrome. J. Neurogastroenterol. Motil. 2018, 24, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Pokusaeva, K.; Johnson, C.; Luk, B.; Uribe, G.; Fu, Y.; Oezguen, N.; Matsunami, R.K.; Lugo, M.; Major, A.; Mori-Akiyama, Y.; et al. GABA-producing Bifidobacterium dentium modulates visceral sensitivity in the intestine. Neurogastroenterol. Motil. 2017, 29. [Google Scholar] [CrossRef] [PubMed]
- Raskov, H.; Burcharth, J.; Pommergaard, H.-C.; Rosenberg, J. Irritable bowel syndrome, the microbiota and the gut-brain axis. Gut Microbes 2016, 7, 365–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terbeck, S.; Savulescu, J.; Chesterman, L.P.; Cowen, P.J. Noradrenaline effects on social behaviour, intergroup relations, and moral decisions. Neurosci. Biobehav. Rev. 2016, 66, 54–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drouin, C.; Bobadilla, A.-C.; Tassin, J.-P. Norepinephrine. In Reference Module in Neuroscience and Biobehavioral Psychology; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar] [CrossRef]
- Naitou, K.; Shiina, T.; Kato, K.; Nakamori, H.; Sano, Y.; Shimizu, Y. Colokinetic effect of noradrenaline in the spinal defecation center: Implication for motility disorders. Sci. Rep. 2015, 5, 12623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kułak-Bejda, A.; Bejda, G.; Waszkiewicz, N. Antidepressants for irritable bowel syndrome—A systematic review. Pharmacol. Rep. 2017, 69, 1366–1379. [Google Scholar] [CrossRef]
- Nozu, T.; Miyagishi, S.; Ishioh, M.; Takakusaki, K.; Okumura, T. Imipramine improves visceral sensation and gut barrier in rat models of irritable bowel syndrome. Eur. J. Pharmacol. 2020, 887, 173565. [Google Scholar] [CrossRef]
- Brinkman, D.J.; ten Hove, A.S.; Vervoordeldonk, M.J.; Luyer, M.D.; de Jonge, W.J. Neuroimmune Interactions in the Gut and Their Significance for Intestinal Immunity. Cells 2019, 8, 670. [Google Scholar] [CrossRef] [Green Version]
- Strandwitz, P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018, 1693, 128–133. [Google Scholar] [CrossRef]
- Kirschstein, T.; Dammann, F.; Klostermann, J.; Rehberg, M.; Tokay, T.; Schubert, R.; Köhling, R. Dopamine induces contraction in the proximal, but relaxation in the distal rat isolated small intestine. Neurosci. Lett. 2009, 465, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Zhang, X.-N.X.-L.; Feng, X.-Y.; Liu, C.-Z.; Zhang, X.-N.X.-L.; Quan, Z.-S.; Yan, J.-T.; Zhu, J.-X. Dopamine promotes colonic mucus secretion through dopamine D5 receptor in rats. Am. J. Physiol. Cell Physiol. 2019, 316, C393–C403. [Google Scholar] [CrossRef] [PubMed]
- Tack, J.; Fried, M.; Houghton, L.A.; Spicak, J.; Fisher, G. Systematic review: The efficacy of treatments for irritable bowel syndrome—A European perspective. Aliment. Pharmacol. Ther. 2006, 24, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Mitsi, V.; Zachariou, V. Modulation of pain, nociception, and analgesia by the brain reward center. Neuroscience 2016, 338, 81–92. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xu, G.; Hu, S.; Wu, H.; Dai, Y.; Zhang, W.; Tang, F.; Luo, H.; Shi, X. Electroacupuncture alleviates intestinal inflammation and barrier dysfunction by activating dopamine in a rat model of intestinal ischaemia. Acupunct. Med. 2020. [Google Scholar] [CrossRef]
- Yan, Y.; Jiang, W.; Liu, L.; Wang, X.; Ding, C.; Tian, Z.; Zhou, R. Dopamine Controls Systemic Inflammation through Inhibition of NLRP3 Inflammasome. Cell 2015, 160, 62–73. [Google Scholar] [CrossRef] [Green Version]
- Weinstein, L.I.; Revuelta, A.; Pando, R.H. Catecholamines and acetylcholine are key regulators of the interaction between microbes and the immune system. Ann. N. Y. Acad. Sci. 2015, 1351, 39–51. [Google Scholar] [CrossRef]
- Hod, K.; Sperber, A.D.; Maharshak, N.; Ron, Y.; Shapira, I.; David, Z.; Rogowski, O.; Berliner, S.; Shenhar-Tsarfaty, S.; Dekel, R. Serum cholinesterase activity is elevated in female diarrhea-predominant irritable bowel syndrome patients compared to matched controls. Neurogastroenterol. Motil. 2018, 30, e13464. [Google Scholar] [CrossRef]
- Burgueño, J.F.; Abreu, M.T. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 263–278. [Google Scholar] [CrossRef]
- de Kivit, S.; Tobin, M.C.; Forsyth, C.B.; Keshavarzian, A.; Landay, A.L. Regulation of Intestinal Immune Responses through TLR Activation: Implications for Pro- and Prebiotics. Front. Immunol. 2014, 5, 60. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Zhang, Q.; Wang, C.; Wu, H.; Jiao, L.; Hong, Q.; Hu, C. LPS challenge increased intestinal permeability, disrupted mitochondrial function and triggered mitophagy of piglets. Innate Immun. 2018, 24, 221–230. [Google Scholar] [CrossRef] [Green Version]
- van Thiel, I.A.M.; Botschuijver, S.; de Jonge, W.J.; Seppen, J. Painful interactions: Microbial compounds and visceral pain. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165534. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, D.; Liston, M.; Hyland, N.P.; Dinan, T.G.; Cryan, J.F. Colonic soluble mediators from the maternal separation model of irritable bowel syndrome activate submucosal neurons via an interleukin-6-dependent mechanism. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G241–G252. [Google Scholar] [CrossRef] [PubMed]
- Brebner, K.; Hayley, S.; Zacharko, R.; Merali, Z.; Anisman, H. Synergistic effects of interleukin-1beta, interleukin-6, and tumor necrosis factor-alpha: Central monoamine, corticosterone, and behavioral variations. Neuropsychopharmacology 2000, 22, 566–580. [Google Scholar] [CrossRef] [Green Version]
- O’Malley, D.; Dinan, T.G.; Cryan, J.F. Interleukin-6 modulates colonic transepithelial ion transport in the stress-sensitive wistar kyoto rat. Front. Pharmacol. 2012, 3, 190. [Google Scholar] [CrossRef] [Green Version]
- Bashashati, M.; Moradi, M.; Sarosiek, I. Interleukin-6 in irritable bowel syndrome: A systematic review and meta-analysis of IL-6 (-G174C) and circulating IL-6 levels. Cytokine 2017, 99, 132–138. [Google Scholar] [CrossRef]
- Martin-Viñas, J.J.; Quigley, E.M.M.M. Immune response in irritable bowel syndrome: A systematic review of systemic and mucosal inflammatory mediators. J. Dig. Dis. 2016, 17, 572–581. [Google Scholar] [CrossRef]
- O’Malley, D.; Buckley, M.M.; McKernan, D.P.; Quigley, E.M.M.; Cryan, J.F.; Dinan, T.G. Soluble mediators in plasma from irritable bowel syndrome patients excite rat submucosal neurons. Brain. Behav. Immun. 2015, 44, 57–67. [Google Scholar] [CrossRef]
- Dal Peraro, M.; van der Goot, F.G. Pore-forming toxins: Ancient, but never really out of fashion. Nat. Rev. Microbiol. 2016, 14, 77–92. [Google Scholar] [CrossRef]
- Chiu, I.M.; Heesters, B.A.; Ghasemlou, N.; Von Hehn, C.A.; Zhao, F.; Tran, J.; Wainger, B.; Strominger, A.; Muralidharan, S.; Horswill, A.R.; et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature 2013, 501, 52–57. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Chen, L.-H.; Xing, C.; Liu, T. Pain regulation by gut microbiota: Molecular mechanisms and therapeutic potential. Br. J. Anaesth. 2019, 123, 637–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, P.J.; Cryan, J.F.; Dinan, T.G.; Clarke, G. Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology 2017, 112, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Simonavicius, N.; Wu, X.; Swaminath, G.; Reagan, J.; Tian, H.; Ling, L. Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J. Biol. Chem. 2006, 281, 22021–22028. [Google Scholar] [CrossRef] [Green Version]
- Metidji, A.; Omenetti, S.; Crotta, S.; Li, Y.; Nye, E.; Ross, E.; Li, V.; Maradana, M.R.; Schiering, C.; Stockinger, B. The Environmental Sensor AHR Protects from Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity 2019, 50, 1542. [Google Scholar] [CrossRef] [PubMed]
- Bock, K.W. Aryl hydrocarbon receptor (AHR) functions: Balancing opposing processes including inflammatory reactions. Biochem. Pharmacol. 2020, 178, 114093. [Google Scholar] [CrossRef]
- Pernomian, L.; Duarte-Silva, M.; de Barros Cardoso, C.R. The Aryl Hydrocarbon Receptor (AHR) as a Potential Target for the Control of Intestinal Inflammation: Insights from an Immune and Bacteria Sensor Receptor. Clin. Rev. Allergy Immunol. 2020, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.A.; Fu, J.; Chang, P.V. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2020, 117, 19376–19387. [Google Scholar] [CrossRef]
- Shimada, Y.; Kinoshita, M.; Harada, K.; Mizutani, M.; Masahata, K.; Kayama, H.; Takeda, K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS ONE 2013, 8, e80604. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-H.J.; Wood, T.K.; Lee, J.-H.J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 2015, 23, 707–718. [Google Scholar] [CrossRef]
- Lomax, A.E.; Pradhananga, S.; Sessenwein, J.L.; O’Malley, D. Bacterial modulation of visceral sensation: Mediators and mechanisms. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G363–G372. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.-R.; Du, L.-J.; He, H.-Q.; Kim, J.J.; Zhao, Y.; Zhang, Y.-W.; Luo, L.; Dai, N. Fructo-oligosaccharide intensifies visceral hypersensitivity and intestinal inflammation in a stress-induced irritable bowel syndrome mouse model. World J. Gastroenterol. 2017, 23, 8321–8333. [Google Scholar] [CrossRef] [PubMed]
- Tarrerias, A.L.; Millecamps, M.; Alloui, A.; Beaughard, C.; Kemeny, J.L.; Bourdu, S.; Bommelaer, G.; Eschalier, A.; Dapoigny, M.; Ardid, D. Short-chain fatty acid enemas fail to decrease colonic hypersensitivity and inflammation in TNBS-induced colonic inflammation in rats. Pain 2002, 100, 91–97. [Google Scholar] [CrossRef]
- Long, X.; Li, M.; Li, L.-X.; Sun, Y.-Y.; Zhang, W.-X.; Zhao, D.-Y.; Li, Y.-Q. Butyrate promotes visceral hypersensitivity in an IBS-like model via enteric glial cell-derived nerve growth factor. Neurogastroenterol. Motil. 2018, 30, e13227. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The Role of Short-Chain Fatty Acids in Health and Disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef]
- Sun, Q.; Jia, Q.; Song, L.; Duan, L. Alterations in fecal short-chain fatty acids in patients with irritable bowel syndrome: A systematic review and meta-analysis. Medicine (Baltimore) 2019, 98, e14513. [Google Scholar] [CrossRef]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Fukumoto, S.; Tatewaki, M.; Yamada, T.; Fujimiya, M.; Mantyh, C.; Voss, M.; Eubanks, S.; Harris, M.; Pappas, T.N.; Takahashi, T. Short-chain fatty acids stimulate colonic transit via intraluminal 5-HT release in rats. Am. J. Physiol. Integr. Comp. Physiol. 2003, 284, R1269–R1276. [Google Scholar] [CrossRef] [Green Version]
- Reigstad, C.S.; Salmonson, C.E.; Rainey, J.F., III; Szurszewski, J.H.; Linden, D.R.; Sonnenburg, J.L.; Farrugia, G.; Kashyap, P.C. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015, 29, 1395–1403. [Google Scholar] [CrossRef] [Green Version]
- Corrêa-Oliveira, R.; Fachi, J.L.; Vieira, A.; Sato, F.T.; Vinolo, M.A.R. Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 2016, 5, e73. [Google Scholar] [CrossRef]
- Vanhoutvin, S.A.L.W.; Troost, F.J.; Kilkens, T.O.C.; Lindsey, P.J.; Hamer, H.M.; Jonkers, D.M.A.E.; Venema, K.; Brummer, R.-J.M. The effects of butyrate enemas on visceral perception in healthy volunteers. Neurogastroenterol. Motil. 2009, 21, 952-e76. [Google Scholar] [CrossRef] [PubMed]
- Nozu, T.; Miyagishi, S.; Nozu, R.; Takakusaki, K.; Okumura, T. Butyrate inhibits visceral allodynia and colonic hyperpermeability in rat models of irritable bowel syndrome. Sci. Rep. 2019, 9, 19603. [Google Scholar] [CrossRef] [PubMed]
- Walters, J.R.F. Bile acid diarrhoea and FGF19: New views on diagnosis, pathogenesis and therapy. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Oduyebo, I.; Camilleri, M. Bile acid disease: The emerging epidemic. Curr. Opin. Gastroenterol. 2017, 33, 189–195. [Google Scholar] [CrossRef] [Green Version]
- Peleman, C.; Camilleri, M.; Busciglio, I.; Burton, D.; Donato, L.; Zinsmeister, A.R. Colonic Transit and Bile Acid Synthesis or Excretion in Patients With Irritable Bowel Syndrome-Diarrhea Without Bile Acid Malabsorption. Clin. Gastroenterol. Hepatol. 2017, 15, 720–727.e1. [Google Scholar] [CrossRef] [Green Version]
- Slattery, S.A.; Niaz, O.; Aziz, Q.; Ford, A.C.; Farmer, A.D. Systematic review with meta-analysis: The prevalence of bile acid malabsorption in the irritable bowel syndrome with diarrhoea. Aliment. Pharmacol. Ther. 2015, 42, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Cangemi, D.J.; Lacy, B.E. Management of irritable bowel syndrome with diarrhea: A review of nonpharmacological and pharmacological interventions. Ther. Adv. Gastroenterol. 2019, 12. [Google Scholar] [CrossRef]
- Zhao, L.; Yang, W.; Chen, Y.; Huang, F.; Lu, L.; Lin, C.; Huang, T.; Ning, Z.; Zhai, L.; Zhong, L.L.; et al. A Clostridia-rich microbiota enhances bile acid excretion in diarrhea-predominant irritable bowel syndrome. J. Clin. Investig. 2020, 130, 438–450. [Google Scholar] [CrossRef] [Green Version]
- Fakhoury, H.M.A.; Kvietys, P.R.; AlKattan, W.; Al Anouti, F.; Elahi, M.A.; Karras, S.N.; Grant, W.B. Vitamin D and intestinal homeostasis: Barrier, microbiota, and immune modulation. J. Steroid Biochem. Mol. Biol. 2020, 200, 105663. [Google Scholar] [CrossRef]
- Clark, A.; Mach, N. Role of Vitamin D in the Hygiene Hypothesis: The Interplay between Vitamin D, Vitamin D Receptors, Gut Microbiota, and Immune Response. Front. Immunol. 2016, 7, 627. [Google Scholar] [CrossRef] [Green Version]
- Hewison, M. Antibacterial effects of vitamin D. Nat. Rev. Endocrinol. 2011, 7, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Zhang, Z.; Musch, M.W.; Ning, G.; Sun, J.; Hart, J.; Bissonnette, M.; Li, Y.C. Novel role of the vitamin D receptor in maintaining the integrity of the intestinal mucosal barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, G208–G216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reich, K.M.; Fedorak, R.N.; Madsen, K.; Kroeker, K.I. Vitamin D improves inflammatory bowel disease outcomes: Basic science and clinical review. World J. Gastroenterol. 2014, 20, 4934–4947. [Google Scholar] [CrossRef] [PubMed]
- Bruewer, M.; Samarin, S.; Nusrat, A. Inflammatory bowel disease and the apical junctional complex. Ann. N. Y. Acad. Sci. 2006, 1072, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-W.; Wang, P.-Y.; Zhu, J.; Chen, G.-W.; Zhang, J.-L.; Chen, Z.-Y.; Zuo, S.; Liu, Y.-C.; Pan, Y.-S. Protective effect of 1,25-dihydroxyvitamin d3 on lipopolysaccharide-induced intestinal epithelial tight junction injury in caco-2 cell monolayers. Inflammation 2015, 38, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhu, J.; Chen, G.; Zuo, S.; Zhang, J.; Chen, Z.; Wang, X.; Li, J.; Liu, Y.; Wang, P. 1,25-Dihydroxyvitamin D3 preserves intestinal epithelial barrier function from TNF-α induced injury via suppression of NF-kB p65 mediated MLCK-P-MLC signaling pathway. Biochem. Biophys. Res. Commun. 2015, 460, 873–878. [Google Scholar] [CrossRef]
- Malaguarnera, L. Vitamin D and microbiota: Two sides of the same coin in the immunomodulatory aspects. Int. Immunopharmacol. 2020, 79, 106112. [Google Scholar] [CrossRef]
- Ligaarden, S.C.; Farup, P.G. Low intake of vitamin B6 is associated with irritable bowel syndrome symptoms. Nutr. Res. 2011, 31, 356–361. [Google Scholar] [CrossRef]
- O’Malley, D. Endocrine regulation of gut function—A role for glucagon-like peptide-1 in the pathophysiology of irritable bowel syndrome. Exp. Physiol. 2019, 104, 3–10. [Google Scholar] [CrossRef]
- Rastelli, M.; Cani, P.D.; Knauf, C. The Gut Microbiome Influences Host Endocrine Functions. Endocr. Rev. 2019, 40, 1271–1284. [Google Scholar] [CrossRef]
- Yang, M.; Fukui, H.; Eda, H.; Xu, X.; Kitayama, Y.; Hara, K.; Kodani, M.; Tomita, T.; Oshima, T.; Watari, J.; et al. Involvement of gut microbiota in association between GLP-1/GLP-1 receptor expression and gastrointestinal motility. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G367–G373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Dwairi, A.; Alqudah, T.E.; Al-Shboul, O.; Alqudah, M.; Mustafa, A.G.; Alfaqih, M.A. Glucagon-like peptide-1 exerts anti-inflammatory effects on mouse colon smooth muscle cells through the cyclic adenosine monophosphate/nuclear factor-κB pathway in vitro. J. Inflamm. Res. 2018, 11, 95–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.-Y.; Zhang, N.; Wen, S.; Zhang, J.; Sun, X.-L.; Fan, X.-M.; Sun, Y.-H. Decreased glucagon-like peptide-1 correlates with abdominal pain in patients with constipation-predominant irritable bowel syndrome. Clin. Res. Hepatol. Gastroenterol. 2017, 41, 459–465. [Google Scholar] [CrossRef] [PubMed]
- El-Salhy, M.; Hatlebakk, J.G.; Hausken, T. Possible role of peptide YY (PYY) in the pathophysiology of irritable bowel syndrome (IBS). Neuropeptides 2020, 79, 101973. [Google Scholar] [CrossRef]
- Ballantyne, G.H. Peptide YY(1-36) and peptide YY(3-36): Part I. Distribution, release and actions. Obes. Surg. 2006, 16, 651–658. [Google Scholar] [CrossRef]
- El-Salhy, M.; Gilja, O.H. Abnormalities in ileal stem, neurogenin 3, and enteroendocrine cells in patients with irritable bowel syndrome. BMC Gastroenterol. 2017, 17, 90. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Zhang, B.; Verne, G.N. Intestinal membrane permeability and hypersensitivity in the irritable bowel syndrome. Pain 2009, 146, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Cheng, P.; Yao, J.; Wang, C.; Zhang, L.; Kong, W. Molecular and cellular mechanisms of tight junction dysfunction in the irritable bowel syndrome. Mol. Med. Rep. 2015, 12, 3257–3264. [Google Scholar] [CrossRef]
- Desormeaux, C.; Bautzova, T.; Garcia-Caraballo, S.; Rolland, C.; Barbaro, M.R.; Brierley, S.M.; Barbara, G.; Vergnolle, N.; Cenac, N. Protease-activated receptor 1 is implicated in irritable bowel syndrome mediators-induced signaling to thoracic human sensory neurons. Pain 2018, 159, 1257–1267. [Google Scholar] [CrossRef]
- Pontarollo, G.; Mann, A.; Brandão, I.; Malinarich, F.; Schöpf, M.; Reinhardt, C. Protease-activated receptor signaling in intestinal permeability regulation. FEBS J. 2020, 287, 645–658. [Google Scholar] [CrossRef] [Green Version]
- Buhner, S.; Hahne, H.; Hartwig, K.; Li, Q.; Vignali, S.; Ostertag, D.; Meng, C.; Hörmannsperger, G.; Braak, B.; Pehl, C.; et al. Protease signaling through protease activated receptor 1 mediate nerve activation by mucosal supernatants from irritable bowel syndrome but not from ulcerative colitis patients. PLoS ONE 2018, 13, e0193943. [Google Scholar] [CrossRef] [PubMed]
- Gecse, K.; Róka, R.; Ferrier, L.; Leveque, M.; Eutamene, H.; Cartier, C.; Ait-Belgnaoui, A.; Rosztóczy, A.; Izbéki, F.; Fioramonti, J.; et al. Increased faecal serine protease activity in diarrhoeic IBS patients: A colonic lumenal factor impairing colonic permeability and sensitivity. Gut 2008, 57, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Edogawa, S.; Edwinson, A.L.; Peters, S.A.; Chikkamenahalli, L.L.; Sundt, W.; Graves, S.; Gurunathan, S.V.; Breen-Lyles, M.; Johnson, S.; Dyer, R.; et al. Serine proteases as luminal mediators of intestinal barrier dysfunction and symptom severity in IBS. Gut 2020, 69, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Róka, R.; Wittmann, T.; Bueno, L. Altered protease signalling in the gut: A novel pathophysiological factor in irritable bowel syndrome. Neurogastroenterol. Motil. 2008, 20, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Barcik, W.; Pugin, B.; Brescó, M.S.; Westermann, P.; Rinaldi, A.; Groeger, D.; Van Elst, D.; Sokolowska, M.; Krawczyk, K.; Frei, R.; et al. Bacterial secretion of histamine within the gut influences immune responses within the lung. Allergy 2019, 74, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; De las Rivas, B.; Marcobal, A.; Muñoz, R. Updated molecular knowledge about histamine biosynthesis by bacteria. Crit. Rev. Food Sci. Nutr. 2008, 48, 697–714. [Google Scholar] [CrossRef] [Green Version]
- Gershon, M.D. 5-Hydroxytryptamine (serotonin) in the gastrointestinal tract. Curr. Opin. Endocrinol. Diabetes Obes. 2013, 20, 14–21. [Google Scholar] [CrossRef] [Green Version]
- Roshchina, V.V. Evolutionary Considerations of Neurotransmitters in Microbial, Plant, and Animal Cells. In Microbial Endocrinology; Springer: New York, NY, USA, 2010; pp. 17–52. [Google Scholar]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [Green Version]
- Tada, Y.; Ishihara, S.; Kawashima, K.; Fukuba, N.; Sonoyama, H.; Kusunoki, R.; Oka, A.; Mishima, Y.; Oshima, N.; Moriyama, I.; et al. Downregulation of serotonin reuptake transporter gene expression in healing colonic mucosa in presence of remaining low-grade inflammation in ulcerative colitis. J. Gastroenterol. Hepatol. 2016, 31, 1443–1452. [Google Scholar] [CrossRef]
- Jin, D.-C.; Cao, H.-L.; Xu, M.-Q.; Wang, S.-N.; Wang, Y.-M.; Yan, F.; Wang, B.-M. Regulation of the serotonin transporter in the pathogenesis of irritable bowel syndrome. World J. Gastroenterol. 2016, 22, 8137–8148. [Google Scholar] [CrossRef]
- Xiao, Q.-Y.; Fang, X.-C.; Li, X.-Q.; Fei, G.-J. Ethnic differences in genetic polymorphism associated with irritable bowel syndrome. World J. Gastroenterol. 2020, 26, 2049–2063. [Google Scholar] [CrossRef] [PubMed]
- Zareian, M.; Ebrahimpour, A.; Bakar, F.A.; Mohamed, A.K.S.; Forghani, B.; Ab-Kadir, M.S.B.; Saari, N. A glutamic acid-producing lactic acid bacteria isolated from Malaysian fermented foods. Int. J. Mol. Sci. 2012, 13, 5482–5497. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, Y.; Hashimoto, K.-I.; Sawada, Y.; Sokabe, M.; Kawasaki, H.; Martinac, B. Corynebacterium glutamicum mechanosensitive channels: Towards unpuzzling “glutamate efflux” for amino acid production. Biophys. Rev. 2018, 10, 1359–1369. [Google Scholar] [CrossRef] [PubMed]
- Barrett, E.; Ross, R.P.; O’Toole, P.W.; Fitzgerald, G.F.; Stanton, C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012, 113, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Asano, Y.; Hiramoto, T.; Nishino, R.; Aiba, Y.; Kimura, T.; Yoshihara, K.; Koga, Y.; Sudo, N. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G1288–G1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishima, Y.; Oka, A.; Liu, B.; Herzog, J.W.; Eun, C.S.; Fan, T.-J.; Bulik-Sullivan, E.; Carroll, I.M.; Hansen, J.J.; Chen, L.; et al. Microbiota maintain colonic homeostasis by activating TLR2/MyD88/PI3K signaling in IL-10-producing regulatory B cells. J. Clin. Investig. 2019, 130. [Google Scholar] [CrossRef] [Green Version]
- Ishihara, S.; Tada, Y.; Fukuba, N.; Oka, A.; Kusunoki, R.; Mishima, Y.; Oshima, N.; Moriyama, I.; Yuki, T.; Kawashima, K.; et al. Pathogenesis of irritable bowel syndrome—Review regarding associated infection and immune activation. Digestion 2013, 87, 204–211. [Google Scholar] [CrossRef]
- Belmonte, L.; Beutheu Youmba, S.; Bertiaux-Vandaële, N.; Antonietti, M.; Lecleire, S.; Zalar, A.; Gourcerol, G.; Leroi, A.-M.; Déchelotte, P.; Coëffier, M.; et al. Role of Toll Like Receptors in Irritable Bowel Syndrome: Differential Mucosal Immune Activation According to the Disease Subtype. PLoS ONE 2012, 7, e42777. [Google Scholar] [CrossRef] [Green Version]
- Brint, E.K.; MacSharry, J.; Fanning, A.; Shanahan, F.; Quigley, E.M.M. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am. J. Gastroenterol. 2011, 106, 329–336. [Google Scholar] [CrossRef]
- Dlugosz, A.; Zakikhany, K.; Acevedo, N.; D’Amato, M.; Lindberg, G. Increased Expression of Toll-Like Receptors 4, 5, and 9 in Small Bowel Mucosa from Patients with Irritable Bowel Syndrome. BioMed Res. Int. 2017, 2017, 9624702. [Google Scholar] [CrossRef]
- Dlugosz, A.; Nowak, P.; D’Amato, M.; Mohammadian Kermani, G.; Nyström, J.; Abdurahman, S.; Lindberg, G. Increased serum levels of lipopolysaccharide and antiflagellin antibodies in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 2015, 27, 1747–1754. [Google Scholar] [CrossRef] [PubMed]
- Villani, A.-C.; Lemire, M.; Thabane, M.; Belisle, A.; Geneau, G.; Garg, A.X.; Clark, W.F.; Moayyedi, P.; Collins, S.M.; Franchimont, D.; et al. Genetic risk factors for post-infectious irritable bowel syndrome following a waterborne outbreak of gastroenteritis. Gastroenterology 2010, 138, 1502–1513. [Google Scholar] [CrossRef] [PubMed]
- Rea, K.; O’Mahony, S.M.; Dinan, T.G.; Cryan, J.F. The Role of the Gastrointestinal Microbiota in Visceral Pain. Handb. Exp. Pharmacol. 2017, 239, 269–287. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.M.; Scott, L.V.; Dinan, T.G. Cytokines: Abnormalities in major depression and implications for pharmacological treatment. Hum. Psychopharmacol. 2004, 19, 397–403. [Google Scholar] [CrossRef]
- Meerveld, B.G.-V.; Johnson, A.C. Mechanisms of Stress-induced Visceral Pain. J. Neurogastroenterol. Motil. 2018, 24, 7–18. [Google Scholar] [CrossRef]
- Akiho, H.; Ihara, E.; Motomura, Y.; Nakamura, K. Cytokine-induced alterations of gastrointestinal motility in gastrointestinal disorders. World J. Gastrointest. Pathophysiol. 2011, 2, 72–81. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Sittipo, P.; Shim, J.-W.; Lee, Y.K. Microbial Metabolites Determine Host Health and the Status of Some Diseases. Int. J. Mol. Sci. 2019, 20, 5296. [Google Scholar] [CrossRef] [Green Version]
- James, S.C.; Fraser, K.; Young, W.; McNabb, W.C.; Roy, N.C. Gut Microbial Metabolites and Biochemical Pathways Involved in Irritable Bowel Syndrome: Effects of Diet and Nutrition on the Microbiome. J. Nutr. 2020, 150, 1012–1021. [Google Scholar] [CrossRef] [Green Version]
- Keszthelyi, D.; Troost, F.J.; Masclee, A.A.M. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol. Motil. 2009, 21, 1239–1249. [Google Scholar] [CrossRef]
- Keszthelyi, D.; Troost, F.J.; Jonkers, D.M.; Kruimel, J.W.; Leue, C.; Masclee, A.A.M. Decreased levels of kynurenic acid in the intestinal mucosa of IBS patients: Relation to serotonin and psychological state. J. Psychosom. Res. 2013, 74, 501–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Zheng, J.; Bai, Y.; Wang, D.; Cui, Z.; Li, Y.; Zhang, J.; Wang, Y. Characterization of kynurenine pathway in patients with diarrhea-predominant irritable bowel syndrome. Eur. J. Histochem. 2020, 64. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tana, C.; Umesaki, Y.; Imaoka, A.; Handa, T.; Kanazawa, M.; Fukudo, S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol. Motil. 2010, 22, 512-e115. [Google Scholar] [CrossRef] [PubMed]
- Kaji, I.; Karaki, S.; Kuwahara, A. Short-chain fatty acid receptor and its contribution to glucagon-like peptide-1 release. Digestion 2014, 89, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, C.B.; Gabe, M.B.N.; Svendsen, B.; Dragsted, L.O.; Rosenkilde, M.M.; Holst, J.J. The impact of short-chain fatty acids on GLP-1 and PYY secretion from the isolated perfused rat colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G53–G65. [Google Scholar] [CrossRef] [Green Version]
- Tolhurst, G.; Heffron, H.; Lam, Y.S.; Parker, H.E.; Habib, A.M.; Diakogiannaki, E.; Cameron, J.; Grosse, J.; Reimann, F.; Gribble, F.M. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 2012, 61, 364–371. [Google Scholar] [CrossRef] [Green Version]
- Wahlström, A.; Sayin, S.I.; Marschall, H.-U.; Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Yang, L.; Wang, Z.; Huang, W. Bile acid nuclear receptor FXR and digestive system diseases. Acta Pharm. Sin. B 2015, 5, 135–144. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Liu, S.; Wang, M.; Hu, H.; Yin, J.; Liu, C.; Huang, Y. Gut Microbiota Dysbiosis Associated with Bile Acid Metabolism in Neonatal Cholestasis Disease. Sci. Rep. 2020, 10, 7686. [Google Scholar] [CrossRef]
- Charoenngam, N.; Shirvani, A.; Kalajian, T.A.; Song, A.; Holick, M.F. The Effect of Various Doses of Oral Vitamin D3 Supplementation on Gut Microbiota in Healthy Adults: A Randomized, Double-blinded, Dose-response Study. Anticancer Res. 2020, 40, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.N.; Cagan, A.; Gainer, V.S.; Cheng, S.-C.; Cai, T.; Szolovits, P.; Shaw, S.Y.; Churchill, S.; Karlson, E.W.; Murphy, S.N.; et al. Higher plasma vitamin D is associated with reduced risk of Clostridium difficile infection in patients with inflammatory bowel diseases. Aliment. Pharmacol. Ther. 2014, 39, 1136–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Liao, A.P.; Xia, Y.; Chun Li, Y.; Li, J.-D.; Sartor, R.B.; Sun, J. Vitamin D Receptor Negatively Regulates Bacterial-Stimulated NF-{kappa}B Activity in Intestine. Am. J. Pathol. 2010, 177, 686–697. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yoon, S.; Zhang, Y.-G.; Lu, R.; Xia, Y.; Wan, J.; Petrof, E.O.; Claud, E.C.; Chen, D.; Sun, J. Vitamin D receptor pathway is required for probiotic protection in colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G341-9. [Google Scholar] [CrossRef] [Green Version]
- Miura, K.; Oshima, T.; Ito, C.; Horikawa, T.; Yamada, M.; Tomita, T.; Fukui, H.; Miwa, H. Vitamin D receptor is overexpressed in the duodenum of patients with irritable bowel syndrome. J. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef]
- El Amrousy, D.; Hassan, S.; El Ashry, H.; Yousef, M.; Hodeib, H. Vitamin D supplementation in adolescents with irritable bowel syndrome: Is it useful? A randomized controlled trial. Saudi J. Gastroenterol. 2018, 24, 109–114. [Google Scholar] [CrossRef]
- Rosenberg, J.; Ischebeck, T.; Commichau, F.M. Vitamin B6 metabolism in microbes and approaches for fermentative production. Biotechnol. Adv. 2017, 35, 31–40. [Google Scholar] [CrossRef]
- Cellini, B.; Zelante, T.; Dindo, M.; Bellet, M.M.; Renga, G.; Romani, L.; Costantini, C. Pyridoxal 5’-Phosphate-Dependent Enzymes at the Crossroads of Host-Microbe Tryptophan Metabolism. Int. J. Mol. Sci. 2020, 21, 5823. [Google Scholar] [CrossRef]
- Huang, S.-C.; Wei, J.C.-C.; Wu, D.J.; Huang, Y.-C. Vitamin B(6) supplementation improves pro-inflammatory responses in patients with rheumatoid arthritis. Eur. J. Clin. Nutr. 2010, 64, 1007–1013. [Google Scholar] [CrossRef] [Green Version]
- El-Salhy, M.; Hausken, T.; Gilja, O.H.; Hatlebakk, J.G. The possible role of gastrointestinal endocrine cells in the pathophysiology of irritable bowel syndrome. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 139–148. [Google Scholar] [CrossRef]
- Gribble, F.M.; Reimann, F. Function and mechanisms of enteroendocrine cells and gut hormones in metabolism. Nat. Rev. Endocrinol. 2019, 15, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vancamelbeke, M.; Vermeire, S. The intestinal barrier: A fundamental role in health and disease. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 821–834. [Google Scholar] [CrossRef] [PubMed]
- König, J.; Wells, J.; Cani, P.D.; García-Ródenas, C.L.; MacDonald, T.; Mercenier, A.; Whyte, J.; Troost, F.; Brummer, R.-J. Human Intestinal Barrier Function in Health and Disease. Clin. Transl. Gastroenterol. 2016, 7, e196. [Google Scholar] [CrossRef] [PubMed]
- Tezuka, H.; Ohteki, T. Regulation of IgA Production by Intestinal Dendritic Cells and Related Cells. Front. Immunol. 2019, 10, 1891. [Google Scholar] [CrossRef] [PubMed]
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef] [PubMed]
- Chung, L.K.; Raffatellu, M.G.I. pros: Antimicrobial defense in the gastrointestinal tract. Semin. Cell Dev. Biol. 2019, 88, 129–137. [Google Scholar] [CrossRef]
- Fan, H.; Wang, A.; Wang, Y.; Sun, Y.; Han, J.; Chen, W.; Wang, S.; Wu, Y.; Lu, Y. Innate Lymphoid Cells: Regulators of Gut Barrier Function and Immune Homeostasis. J. Immunol. Res. 2019, 2019, 2525984. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, L.; Qin, S. Gut Microbiota Modulation on Intestinal Mucosal Adaptive Immunity. J. Immunol. Res. 2019, 2019, 4735040. [Google Scholar] [CrossRef]
- Hyland, N.P.; Quigley, E.M.M.; Brint, E. Microbiota-host interactions in irritable bowel syndrome: Epithelial barrier, immune regulation and brain-gut interactions. World J. Gastroenterol. 2014, 20, 8859–8866. [Google Scholar] [CrossRef]
- Steck, N.; Mueller, K.; Schemann, M.; Haller, D. Bacterial proteases in IBD and IBS. Gut 2012, 61, 1610–1618. [Google Scholar] [CrossRef] [PubMed]


| Microbial Products | Visceral Pain | GI Motility | Mucosal Permeability | Mucosal Inflammation | Influencing Dysbiosis | References |
|---|---|---|---|---|---|---|
| Neurotransmitters | ||||||
| Histamine | ↑↑ | ↑ | ↑ | ↑ | [53,78,79,80,81,82,83,84] | |
| Serotonin | ↑ | ↑ | ↑ | ↑ | ++ | [7,85,86,87,88] |
| Glutamate | ↑↑ | ↓? | ↑ | + | [89,90,91,92,93,94,95,96,97,98,99] | |
| γ-aminobutyric acid | ↓ | ↓ | ↓ | [92,100,101,102,103,104,105] | ||
| Noradrenalin | ↑ | ↑ | ↑ | ↑↓ | [106,107,108,109,110,111] | |
| Dopamine | ↓ | ↑↓ | ↑? | ↓? | + | [112,113,114,115,116,117,118] |
| Acetylcholine | ↑ | ↓ | [111,112,119,120] | |||
| Compounds | ||||||
| Toll-like receptor ligands | ↑↓ | ↑↓ | ↑↓ | ↑↓ | [121,122,123] | |
| Cytokines | ↑↓ | ↑↓ | ↑↓ | ↑↓ | [52,124,125,126,127,128,129,130] | |
| Pore-forming toxins, N-formylated peptides | ↑ | ↑ | ↑ | [124,131,132,133] | ||
| Metabolites | ||||||
| Tryptophan (aryl hydrocarbon receptor, kynurenine pathways) | ↓↑ | ↓ | ↓ | + | [89,124,134,135,136,137,138,139,140,141] | |
| Short-chain fatty acids | ↑↓ | ↑ | ↓ | ↓ | [70,142,143,144,145,146,147,148,149,150,151,152,153,154] | |
| Bile acids | ↓ | ↑ | ↑ | + | [155,156,157,158,159,160] | |
| Vitamin D and B6 | ↓ | ↓ | ↓ | + | [161,162,163,164,165,166,167,168,169,170] | |
| Endocrine factors | ||||||
| Glucagon-like peptide-1 | ↓ | ↓ | ↓ | [171,172,173,174,175] | ||
| Peptide YY | ↓ | ↓ | [172,176,177,178] | |||
| Enzymes | ||||||
| Proteases | ↑ | ↑↓ | ↑↑↑ | ↑ | [124,179,180,181,182,183,184,185,186] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishima, Y.; Ishihara, S. Molecular Mechanisms of Microbiota-Mediated Pathology in Irritable Bowel Syndrome. Int. J. Mol. Sci. 2020, 21, 8664. https://doi.org/10.3390/ijms21228664
Mishima Y, Ishihara S. Molecular Mechanisms of Microbiota-Mediated Pathology in Irritable Bowel Syndrome. International Journal of Molecular Sciences. 2020; 21(22):8664. https://doi.org/10.3390/ijms21228664
Chicago/Turabian StyleMishima, Yoshiyuki, and Shunji Ishihara. 2020. "Molecular Mechanisms of Microbiota-Mediated Pathology in Irritable Bowel Syndrome" International Journal of Molecular Sciences 21, no. 22: 8664. https://doi.org/10.3390/ijms21228664